Abstract
Background
It is thought that overexpression of epidermal growth factor receptor (EGFR) in non‐small cell lung cancer (NSCLC) might compromise patient survival, presumably by promoting tumour growth by an autocrine mechanism. However, conflicting results have been reported from various laboratories, and the clinical importance of EGFR overexpression remains unsettled.
Methods
A meta‐analysis of previous studies was performed to quantitatively review the effects of EGFR overexpression on survival in patients with NSCLC using a DerSimonian‐Laird random effects model. Eighteen studies including 2972 patients were subjected to final analysis.
Results
Overall, positivity for EGFR overexpression differed between histological types: 39% in adenocarcinomas, 58% in squamous cell carcinomas, 38% in large cell carcinomas, and 32% in cancers in a miscellaneous category (p<0.0001). The combined hazard ratio (HR) was 1.14 (95% CI 0.97 to 1.34; p = 0.103), indicating that EGFR overexpression has no significant impact on survival. When only the 15 immunohistochemistry based studies were considered, the combined HR was 1.08 (95% CI 0.92 to 1.28; p = 0.356), again suggesting that EGFR overexpression has no impact on survival. Heterogeneity testing indicated that there was heterogeneity between studies but publication bias was absent, which suggests that the summary statistics obtained may approximate the actual average.
Conclusions
EGFR overexpression was not associated with poorer survival in patients with NSCLC. Specific mutations of the EGFR gene will need further study in terms of survival implications.
Keywords: epidermal growth factor receptor, lung cancer, overexpression, survival
Lung cancer is the deadliest cancer worldwide.1 Despite tremendous efforts to reduce deaths due to lung cancer, the overall 5 year survival rate is approximately 10% in every country and has shown little improvement in recent decades.2 For earlier disease stages of non‐small cell lung cancer (NSCLC), surgery is potentially curative while, for more advanced stages, chemotherapy and radiotherapy are regarded as standard treatments. Recently a new strategy—molecular targeted therapy—has been considered as a possible addition to this armamentarium. A prototypical molecular target for clinical application is the epidermal growth factor receptor (EGFR).3 Activation of EGFR results in enhancement of cell proliferation and suppression of apoptosis.4,5,6 EGFR therefore seems a logical choice of target for controlling tumour growth.
Among the various anti‐EGFR agents, gefitinib7,8 and erlotinib,9 both tyrosine kinase inhibitors, have proved useful in patients with advanced NSCLC. In particular, gefitinib is now used as a second or third line treatment after platinum‐based chemotherapy, obtaining responses in certain tumours showing resistance to other chemotherapy. In other clinical trials cetuximab, a monoclonal antibody that blocks EGFR, has been confirmed to have antitumour effects in patients with advanced NSCLC.10 The importance of EGFR as a molecular target has therefore been rapidly increasing.
Overexpression of EGFR is thought to imply poor survival, presumably reflecting autocrine stimulation of tumour growth initiated by interaction between EGFR and its ligands, epidermal growth factor (EGF)/transforming growth factor alpha (TGFα), amphiregulin, epiregulin, and beta‐cellulin.11 The association between EGFR overexpression and altered survival in patients with NSCLC has been studied for many years as a test of this hypothesis. However, conflicting results have been reported from different laboratories. Accurate and conclusive evaluation of the clinical significance of EGFR overexpression is important to understand the pharmacological mechanism of anti‐EGFR therapy and to choose appropriate future molecular targets. We therefore carried out a meta‐analysis of published studies to quantitatively review the effects of EGFR overexpression on survival in patients with NSCLC.
Methods
Eligibility criteria for meta‐analysis
This meta‐analysis was limited to studies dealing with the prognostic implications of EGFR overexpression. The following criteria for eligibility among studies were set before collecting articles: (1) Expression of the EGFR gene was evaluated in primary lung cancer tissue as opposed to sera or metastatic tissue. (2) Methods used to evaluate EGFR expression included immunohistochemistry (IHC) or quantification of EGFR mRNA or protein. (3) The histological type of the tumours was NSCLC. (4) Hazard ratio (HR) and its confidence interval (CI) comparing patients with EGFR overexpression with patients without overexpression were described or statistically extractable from the data in the article. (5) Median follow up time exceeded 2 years. (6) Articles were published in English in the periodical literature from January 1990 to November 2004. (7) When multiple articles were published by the same authors or group, the newest or most informative single article was selected.
Collection of published studies
The MEDLINE database was searched online in November 2004 for bibliographic information concerning articles about EGFR expression status and survival in patients with NSCLC. Key words “lung cancer + epidermal growth factor receptor + prognosis”, “lung cancer + epidermal growth factor + survival”, “lung cancer + HER1”, and “lung cancer + erbB1” hit 127, 104, 17, and 20 citations, respectively. Manual selection of relevant studies was carried out based on the summary analysis. Articles found to overlap others or to be unrelated to our question were excluded. When appropriate, items from hand searched bibliographies were added. Thirty seven articles concerning association of EGFR expression with survival in NSCLC were initially found. Among these selected articles, HR and its CI were not extractable statistically from 10 articles,12,13,14,15,16,17,18,19,20,21 which typically reported only p values. Five articles22,23,24,25,26 originated from investigative groups already represented. In three articles27,28,29 EGFR protein was measured in serum. One article30 compared a combined EGFR overexpression or underexpression group with a medium expression group. These 19 papers were excluded from the meta‐analysis (table 1), leaving 18 studies31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48 which fulfilled the eligibility criteria (table 2).
Table 1 Articles excluded from the meta‐analysis.
| Author | Year | Reasons for exclusion | Histology: no of patients | EGFR overexpression(%) | EGFR effect on survival |
|---|---|---|---|---|---|
| Dazzi et al12 | 1989 | No statistical data for extracting HR | Ad: 31 | 49 | NS |
| Sq: 97 | |||||
| La: 7 | |||||
| Mis: 17 | |||||
| Rusch et al22 | 1993 | Updated by Rusch et al34 | Sq: 19 | 45 | NS |
| Non‐sq: 25 | |||||
| Volm et al23 | 1993 | Updated by Volm et al37 | Total: 121 | 83 | S |
| Giatromanolaki et al13 | 1996 | No statistical data for extracting HR | Ad: 38 | 79 | NS |
| Sq: 69 | |||||
| Giatromanolaki et al14 | 1996 | No statistical data for extracting HR | Ad: 38 | 79 | NS |
| Sq: 69 | |||||
| Pfeiffer et al24 | 1996 | Updated by Pfeiffer et al36 | Ad: 59 | 55 | NS |
| Sq: 102 | |||||
| La: 25 | |||||
| Koukourakis et al15 | 1997 | No statistical data for extracting HR | Ad: 38 | ND | NS |
| Sq: 69 | |||||
| Greatens et al16 | 1998 | No statistical data for extracting HR | Ad: 51 | 65 | NS |
| Sq: 44 | |||||
| La: 6 | |||||
| D'Amico et al17 | 2000 | No statistical data for extracting HR | Ad: 208 | 52 | NS |
| Sq: 148 | |||||
| La: 52 | |||||
| Cox et al25 | 2001 | Updated by Swinson et al48 | Ad: 50 | 56 | NS |
| Sq: 105 | |||||
| La: 14 | |||||
| Volm et al18 | 2002 | No statistical data for extracting HR | Ad: 59 | ND | S |
| Sq: 123 | |||||
| La: 34 | |||||
| Kanematsu et al19 | 2003 | No statistical data for extracting HR | Sq: 14 | 81 | NS |
| Non‐sq: 22 | |||||
| Sasaki et al27 | 2003 | Serum protein measured | Ad: 51 | 34 | NS |
| Sq: 42 | |||||
| La: 5 | |||||
| Mis: 9 | |||||
| Brattstrom et al20 | 2004 | No statistical data for extracting HR | Ad: 25 | 30 | NS |
| Sq: 22 | |||||
| La: 2 | |||||
| Mis: 4 | |||||
| Gregore et al28 | 2004 | Serum protein measured | Ad: 29 | ND | NS |
| Non‐ad: 17 | |||||
| Jacot et al29 | 2004 | Serum protein measured | Ad: 56 | ND | NS |
| Sq: 127 | |||||
| La: 38 | |||||
| Niemiec et al30 | 2004 | Over‐ and underexpression compared with medium expression | Sq: 78 | 60 | S |
| Swinson et al26 | 2004 | Updated by Swinson et al48 | Ad: 49 | ND | NS |
| Sq: 107 | |||||
| La: 12 | |||||
| Mis: 4 | |||||
| Ad: 56 | |||||
| Onn et al21 | 2004 | No statistical data for extracting HR | Sq: 40 | 50 | NS |
| Mis: 15 |
EGFR, epidermal growth factor receptor; HR, hazard ratio; Ad, adenocarcinoma; Sq, squamous cell carcinoma; La, large cell carcinoma; Mis, miscellaneous; ND, no data; S, significant; NS, not significant; CFS, cancer free survival.
Table 2 Studies included in the present meta‐analysis.
| Author | Year | Method | Specimen | Antibody | Stage | Histology: no of patients | EGFR overexpression(%) | EGFR effect on survival |
|---|---|---|---|---|---|---|---|---|
| Veale et al31 | 1993 | Protein assay (RIA) | Frozen | NA | I–III | Ad: 7 | 11/19 (58) | S (worse) |
| Sq: 10 | ||||||||
| La: 2 | ||||||||
| Tateishi et al32 | 1994 | IHC | Paraffin | Mouse sera(Transformation Research Inc) | I–IV | Ad: 119 | 55/119 (46) | NS |
| Pastorino et al33 | 1997 | IHC | Paraffin | Monoclonal (Triton Diagnostics) | I | Ad: 217 | 245/515 (48) | NS |
| Sq: 252 | ||||||||
| Mis: 46 | ||||||||
| Rusch et al34 | 1997 | IHC | Paraffin | Monoclonal (Triton Diagnostics) | I–IIIA | Ad: 52 | 68/96 (71) | S (better) |
| Sq: 36 | ||||||||
| La: 8 | ||||||||
| Fontanini et al35 | 1998 | IHC | Paraffin | Monoclonal (Triton Diagnostics) | I–IIIA | Ad: 66 | 92/195 (47) | NS |
| Sq: 116 | ||||||||
| Mis: 13 | ||||||||
| Pfeiffer et al36 | 1998 | ELISA | Frozen | (Amersham, Oncogene Science) | ND | Ad: 57 | 49/180 (27) | NS |
| Sq: 100 | ||||||||
| La: 23 | ||||||||
| Volm et al37 | 1998 | IHC | Paraffin | Polyclonal (Dianova) | I–III | Sq: 121 | 100/121 (83) | S (worse) |
| D'Amico et al38 | 1999 | IHC | Paraffin | Monoclonal (BioGenex Laboratories) | I | Ad: 208 | 212/408 (52) | NS |
| Sq: 145 | ||||||||
| Mis: 55 | ||||||||
| Fu et al39 | 1999 | IHC | Paraffin | Monoclonal(Sigma) | I–IIIB | Sq: 63 | 104/158 (66) | NS |
| Non‐sq: 95 | ||||||||
| Ohsaki et al40 | 2000 | IHC | Paraffin | Monoclonal(Novacastra) | I–IV | Ad: 142 | ||
| Sq: 127 | 124/290 (43) | S (worse) | ||||||
| La: 17 | ||||||||
| Mis: 4 | ||||||||
| Brabender et al41 | 2001 | RT‐PCR | Frozen | NA | I–IIIA | Ad: 32 | 28/83 (34) | NS |
| Sq: 39 | ||||||||
| La: 12 | ||||||||
| Piyathilake et al42 | 2002 | IHC | Paraffin | Monoclonal(Zymed Laboratories) | I–IIIA | Sq: 60 | 30/60 (50)* | S (worse) |
| Hirsch et al43 | 2003 | IHC | Paraffin | Monoclonal(Zymed Laboratories) | I–IIIB | Ad: 79 | 68/183 (37) | NS |
| Sq: 89 | ||||||||
| La: 15 | ||||||||
| Deeb et al44 | 2004 | IHC | Paraffin | Monoclonal(Dako) | I–IIIA | Ad: 83 | 43/123 (35) | NS |
| Sq: 43 | ||||||||
| Parra et al45 | 2004 | IHC | Paraffin | Monoclonal(EFeAb‐10) | I–IV | Ad: 29 | 23/50† (46) | S (worse) |
| Sq: 9 | ||||||||
| Mis: 12 | ||||||||
| Selvaggi et al46 | 2004 | IHC | Paraffin | Monoclonal(Oncogene Science) | I–IIIA | Ad: 48 | 48/130 (37) | S (worse) |
| Sq: 60 | ||||||||
| La: 22 | ||||||||
| Shah et al47 | 2004 | IHC | Paraffin | Monoclonal(ZymedLaboratories) | I–III | Ad: 43 | 48/63 (76) | S (better) |
| Sq: 9 | ||||||||
| Mis: 11 | ||||||||
| Swinson et al48 | 2004 | IHC | Paraffin | Monoclonal(Novacastra) | I–IIIA | ND | 93/179 (52) | NS |
EGFR, epidermal growth factor receptor; IHC, immunohistochemistry; ELISA, enzyme linked immunosorbent assay; RTPCR, reverse transcription‐polymerase chain reaction; Ad, adenocarcinoma; Sq, squamous cell carcinoma; La, large cell carcinoma; Mis, miscellaneous; S, significant; NS, not significant; NA, not applicable; ND, no data.
*Median used for cut off value.
†Treated with gefitinib.
Extraction of hazard ratios
HRs and their 95% CIs were used to combine the data. When described in text or tables, we obtained these values directly from six articles.31,33,44,46,47,48 When these statistical variables were not given explicitly in an article, they were calculated from available numerical data in three articles35,38,39 using methods reported by Parmar et al.49 In summary, HRs and CIs were calculated from differences between numbers of observed and expected events concerning the primary end point of length of survival. Published survival curves were used to obtain HRs and CIs in nine articles.32,34,36,37,40,41,42,43,45 In these cases, after dividing the time axis into non‐overlapping intervals, log HR and its variance for each interval were calculated. These estimated values were combined in a stratified manner to obtain the overall HR and 95% CI.49
Statistical analyses
A DerSimonian‐Laird random effects analysis50 was used to estimate the effects of EGFR overexpression on survival. The DerSimonian‐Laird method is the simplest form of a random effects meta‐analysis. To undertake a random effects meta‐analysis, the standard errors of the study‐specific estimates are adjusted to incorporate a measure of the extent of variation (or heterogeneity) among the treatment effects observed in different studies. The size of this adjustment can be estimated from the treatment effects and standard errors of the studies included in the meta‐analysis. This method permits combined assessment of heterogeneous studies to produce combined HR and 95% CI values. Heterogeneity among combined studies was tested simultaneously. The statistical software used was StatsDirect Version 2.3.8. Categorical variables such as EGFR positivity for each histological type were tested by the χ2 test. For these analyses, a p value below 0.05 was considered to indicate significance. Evidence of publication bias was sought by the methods of Egger et al51 and of Begg and Mazumdar.52 For these analyses, p values below 0.1 were considered to indicate significance.
Results
A total of 2972 patients were subjected to final analysis (mean 165 per study; range 19–515). Patients with locoregional disease (stages I–IIIB) were included in 15 studies while patients in stage IV were included in three studies. Surgery was performed for most patients. Only 50 patients in one study45 were treated with gefitinib. Among the 18 studies included in this meta‐analysis, a significant association between EGFR overexpression and survival was found in eight (44.4%), including six (33.3%) linking EGFR expression with shorter survival and two (11.1%) linking EGFR expression with longer survival. The remaining 10 studies (55.6%) yielded negative results.
In the 18 studies included in the analysis, expression of EGFR was evaluated by IHC in 15, by mRNA expression in one, by radioimmunoassay (RIA) in one, and by enzyme linked immunosorbent assay (ELISA) in one. Using cut off values for overexpression chosen by each author, 1441 (48.5%) of the 2972 patients in the meta‐analysis had EGFR positive lung cancers. Only adenocarcinomas were examined in one study and only squamous cell carcinomas were analysed in three. The remaining 14 studies investigated all histological types, although precise numbers of cases for each histological type were not mentioned in three studies.31,39,48 Positivity for EGFR overexpression in the individual studies ranged from 27%36 to 83%.37 Overall EGFR positivity in the studies included differed according to histological type: 39% in adenocarcinomas, 58% in squamous cell carcinomas, 38% in large cell carcinomas, and 32% in miscellaneous lung cancer (table 3, p<0.0001). Overexpression was therefore significantly more frequent in squamous cell carcinoma.
Table 3 Overall numbers and percentages of EGFR positivity according to histological type in cases from included studies.
| Histological type | EGFR positive | p value |
|---|---|---|
| Ad | 460/1175 (39.1%) | <0.0001* |
| Sq | 695/1206 (57.6%) | |
| La | 37/98 (37.8%) | |
| Mis | 41/130 (31.5%) | |
| Total | 1233/2609 (45.8%)† |
EGFR, epidermal growth factor receptor; Ad, adenocarcinoma; Sq, squamous cell carcinoma; La, large cell carcinoma; Mis, miscellaneous.
*χ2 test.
†Three studies were excluded because they lacked information about EGFR positivity according to histological type.
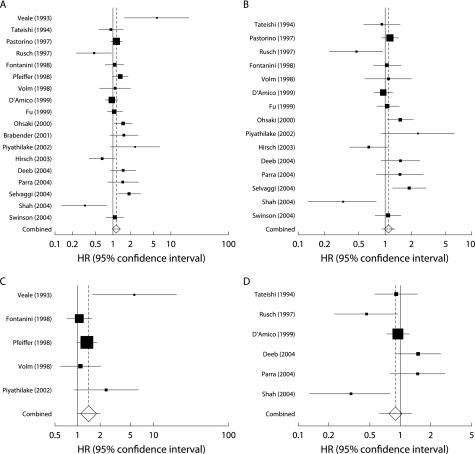
The combined HR for all 18 eligible studies was 1.14 (95% CI 0.97 to 1.34; p = 0.103), indicating that EGFR overexpression had no significant survival impact in patients with NSCLC (fig 1A). We also separately analysed studies using only IHC for evaluation. In these 15 studies the combined HR was 1.08 (95% CI 0.92 to 1.28; p = 0.356), again suggesting that EGFR overexpression had no significant impact on survival (fig 1B). Heterogeneity testing revealed that both the group including all studies and the group including only IHC studies were heterogeneous (Q = 40.8, p = 0.001 for all eligible studies and Q = 31.9, p = 0.004 for IHC studies). We then focused on the predominant histological type in each study. When we limited the analysis to the five studies dealing mostly (>50%) with squamous cell carcinomas, the combined HR was 1.40 (95% CI 0.98 to 2.00; p = 0.07) without heterogeneity (Q = 8.13, p = 0.09; fig 1C). When we limited the analysis to the six studies dealing mostly (>50%) with adenocarcinomas, the combined HR was 0.90 (95% CI 0.63 to 1.28; p = 0.55) with inter‐study heterogeneity (Q = 14.2, p = 0.01; fig 1D).
Figure 1 Meta‐analysis of hazard ratio (HR) of the effect of epidermal growth factor receptor (EGFR) overexpression on survival in patients with non‐small cell lung cancer (NSCLC). Bars indicate 95% confidence interval (CI) of HR in patients with EGFR overexpression. Areas of squares are proportional to weights used in combining data. The centre of the lozenge gives the combined HR. The effect of EGFR on survival was considered statistically significant if the 95% CI for combined HR did not overlap the value 1. (A) All 18 eligible studies; (B) 15 immunohistochemically evaluated studies; (C) five studies including over 50% squamous cell carcinoma cases; and (D) six studies including over 50% adenocarcinoma cases.
Publication bias statistics obtained by the methods of Egger et al51 and Begg and Mazumdar52 were as follows: all 18 eligible studies (p = 0.715 and p = 0.654); 15 IHC studies (p = 0.684 and p = 0.697); five studies considering mainly squamous cell carcinoma (p = 0.153 and p = 0.233); and six studies considering mainly adenocarcinoma (p = 0.525 and p = 0.136). This suggested absence of publication bias in all studies.
Discussion
Gene amplification or overexpression of EGFR in lung cancer has been reported since the 1980s.53,54 EGFR has recently attracted attention as a molecular target in the treatment of NSCLC. Understanding the mechanisms of action of anti‐EGFR therapy requires correct evaluation of the impact of EGFR expression on patient survival. Combining 18 published studies which included 2972 patients with NSCLC yielded summary statistics indicating that EGFR overexpression has no significant impact on survival.
Two major types of meta‐analysis may be performed: meta‐analysis of the literature (MAL) and meta‐analysis of individual patient data (MAP).55 When individual patient data can be accessed easily, MAP is preferable for more precise statistical evaluation. However, we used MAL because individual patient data were difficult to access in the 18 different studies published over a 10 year period.
Meta‐analysis was originally developed to combine the results of randomised controlled trials.56 Recently, however, this approach has been applied successfully to identification of prognostic indicators in patients with lung cancer such as aneuploidy57 and p53 mutations.58 One previous meta‐analysis59 found an association between EGFR overexpression detected by IHC and poor survival in patients with NSCLC. That analysis showed significantly shorter survival based on eight IHC studies (HR 1.13, 95% CI 1.00 to 1.28). However, in that analysis one IHC study34 showing longer survival in tumours overexpressing EGFR was excluded, which might have affected the results. Our present analysis of 15 IHC studies suggests that EGFR overexpression has no effect on survival.
Heterogeneity testing detected significant heterogeneity between the studies in our analysis; even when we limited the scope of analysis to IHC studies only, heterogeneity was still detected. Since EGFR positivity may be related to histological type, we initially thought that heterogeneity may have arisen from the squamous cell carcinomas included. When analysis was limited to the five studies including mostly squamous cell carcinomas, heterogeneity disappeared. On the other hand, when we analysed only the six studies including mostly adenocarcinomas, heterogeneity was still detected. Histological type may therefore be neither a major cause of heterogeneity nor a decisive factor in the biological effect of EGFR overexpression.
Factors related to immunostaining can also cause heterogeneity. Atkins et al60 studied variables affecting the results of immunostaining for EGFR in paraffin embedded specimens and concluded that fixation solution and prolonged storage of archival tissue section might affect the positivity of cells. Other differences concerning staining techniques and evaluation criteria for EGFR positivity might result in heterogeneity between studies.
Publication bias61 is a well known problem in meta‐analysis; positive results tend to be accepted by journals while negative results are often rejected or not even submitted.62 However, our analysis did not suggest publication bias, so the summary statistics obtained may approximate the actual average.
In our meta‐analysis, when HR and CI were not described in the reports they were extracted from the data according to the method of Parmar et al.49 We should keep in mind that estimated HR values and their CIs may differ to some extent depending on the estimation method used.
Theoretically, gefitinib might be most effective for squamous cell carcinomas which express ample EGFR in more than half of cases. However, data obtained to date63,64 suggest that the most effective response may be observed in adenocarcinomas. This discrepancy may require a considerable amount of work to be resolved. In recent studies,65,66,67 adenocarcinomas that showed missense substitutions or small in‐frame deletions within the coding region of the ATP binding pocket in the EGFR gene responded well to treatment with gefitinib. The mutant EGFRs may selectively activate Akt and signal transduction and activator of transcription (STAT) which strongly suppress apoptosis.5 Gefitinib induces apoptosis by preventing mutant EGFR from activating these pathways.
In contrast to the above, Kim et al68 reported that overexpression of EGFR in adenocarcinoma did not predict a response to gefitinib. Furthermore, Perez‐Soler et al69 found that overexpression of EGFR was not a significant predictor for either tumour response to erlotinib or survival after such treatment. In combination, these results and our own suggest that EGFR mutations rather than overexpression are likely to be important determinants of the biological behaviour of NSCLC.
In conclusion, EGFR overexpression was not associated with survival in patients with NSCLC in the present meta‐analysis. Although overexpression was frequently detected in squamous cell carcinoma, there was no effect on survival when studies were selected according to the percentages of squamous cell carcinoma or adenocarcinoma. Since somatic mutations within the EGFR gene are known to result in inhibition of apoptosis, mutations of this gene will require further study in terms of survival implications for patients with NSCLC.
Abbreviations
EGFR - epidermal growth factor receptor
HR - hazard ratio
NSCLC - non‐small cell lung cancer
Footnotes
This study was funded in part by grant no. 15591493 from the Ministry of Education, Science, Sports, and Culture of Japan.
Competing interests: none declared.
References
- 1.Alberg A J, Samet J M. Epidemiology of lung cancer. Chest 200312321–49S. [DOI] [PubMed] [Google Scholar]
- 2.Janssen‐Heijnen M L, Coebergh J W. The changing epidemiology of lung cancer in Europe. Lung Cancer 200341245–258. [DOI] [PubMed] [Google Scholar]
- 3.Dancey J E, Freidlin B. Targeting epidermal growth factor receptor: Are we missing the mark? Lancet 200336262–64. [DOI] [PubMed] [Google Scholar]
- 4.Scagliotti G V, Selvaggi G, Novello S.et al The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res 2004104227–32S. [DOI] [PubMed] [Google Scholar]
- 5.Sordella R, Bell D W, Haber D A.et al Gefitinib‐sensitizing EGFR mutations in lung cancer activate anti‐apoptotic pathways. Science 20043051163–1167. [DOI] [PubMed] [Google Scholar]
- 6.Mukohara T, Kudoh S, Matsuura K.et al Activated Akt expression has significant correlation with EGFR and TGF‐alpha expressions in stage I NSCLC. Anticancer Res 20042411–17. [PubMed] [Google Scholar]
- 7.Johnson D H. Gefitinib (Iressa) trials in non‐small cell lung cancer. Lung Cancer 200341(Suppl 1)S23–S28. [DOI] [PubMed] [Google Scholar]
- 8.Fukuoka M, Yano S, Giaccone G.et al Multi‐institutional randomized phase II trial of gefitinib for previously treated patients with advanced non‐small‐cell lung cancer. J Clin Oncol 2003212237–2246. [DOI] [PubMed] [Google Scholar]
- 9.Perez‐Soler R. The role of erlotinib (Tarceva, OSI 774) in the treatment of non‐small cell lung cancer. Clin Cancer Res 2004104238–40S. [DOI] [PubMed] [Google Scholar]
- 10.Govindan R. Cetuximab in advanced non‐small cell lung cancer. Clin Cancer Res 2004104241–4S. [DOI] [PubMed] [Google Scholar]
- 11.Park J Y, Su Y Q, Ariga M.et al EGF‐like growth factors as mediators of LH action in the ovulatory follicle. Science 2004303682–684. [DOI] [PubMed] [Google Scholar]
- 12.Dazzi H, Hasleton P S, Thatcher N.et al Expression of epidermal growth factor receptor (EGF‐R) in non‐small cell lung cancer. Use of archival tissue and correlation of EGF‐R with histology, tumour size, node status and survival. Br J Cancer 198959746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giatromanolaki A, Koukourakis M I, O'Byrne K.et al Non‐small cell lung cancer: c‐erbB‐2 overexpression correlates with low angiogenesis and poor prognosis. Anticancer Res 1996163819–3825. [PubMed] [Google Scholar]
- 14.Giatromanolaki A, Koukourakis M I, O'Byrne K.et al Prognostic value of angiogenesis in operable non‐small cell lung cancer. J Pathol 199617980–88. [DOI] [PubMed] [Google Scholar]
- 15.Koukourakis M I, Giatromanolaki A, O'Byrne K J.et al Potential role of bcl‐2 as a suppressor of tumour angiogenesis in non‐small‐cell lung cancer. Int J Cancer 199774565–570. [DOI] [PubMed] [Google Scholar]
- 16.Greatens T M, Niehans G A, Rubins J B.et al Do molecular markers predict survival in non‐small‐cell lung cancer? Am J Respir Crit Care Med 19981571093–1097. [DOI] [PubMed] [Google Scholar]
- 17.D'Amico T A, Aloia T A, Moore M B.et al Molecular biologic substaging of stage I lung cancer according to gender and histology. Ann Thorac Surg 200069882–886. [DOI] [PubMed] [Google Scholar]
- 18.Volm M, Koomagi R, Mattern J.et al Expression profile of genes in non‐small cell lung carcinomas from long‐term surviving patients. Clin Cancer Res 200281843–1848. [PubMed] [Google Scholar]
- 19.Kanematsu T, Yano S, Uehara H.et al Phosphorylation, but not overexpression, of epidermal growth factor receptor is associated with poor prognosis of non‐small cell lung cancer patients. Oncol Res 200313289–298. [DOI] [PubMed] [Google Scholar]
- 20.Brattstrom D, Wester K, Bergqvist M.et al HER‐2, EGFR, COX‐2 expression status correlated to microvessel density and survival in resected non‐small cell lung cancer. Acta Oncol 20044380–86. [DOI] [PubMed] [Google Scholar]
- 21.Onn A, Correa A M, Gilcrease M.et al Synchronous overexpression of epidermal growth factor receptor and HER2‐neu protein is a predictor of poor outcome in patients with stage I non‐small cell lung cancer. Clin Cancer Res 200410136–143. [DOI] [PubMed] [Google Scholar]
- 22.Rusch V, Baselga J, Cordon‐Cardo C.et al Differential expression of the epidermal growth factor receptor and its ligands in primary non‐small cell lung cancers and adjacent benign lung. Cancer Res 1993532379–2385. [PubMed] [Google Scholar]
- 23.Volm M, Drings P, Wodrich W. Prognostic significance of the expression of c‐fos, c‐jun and c‐erbB‐1 oncogene products in human squamous cell lung carcinomas. J Cancer Res Clin Oncol 1993119507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiffer P, Clausen P P, Andersen K.et al Lack of prognostic significance of epidermal growth factor receptor and the oncoprotein p185HER‐2 in patients with systemically untreated non‐small‐cell lung cancer: an immunohistochemical study on cryosections. Br J Cancer 19967486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox G, Jones J L, Audi A.et al A biological model for operable non‐small cell lung cancer. Thorax 200156561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swinson D E, Jones J L, Cox G.et al Hypoxia‐inducible factor‐1 alpha in non small cell lung cancer: relation to growth factor, protease and apoptosis pathways. Int J Cancer 200411143–50. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki H, Yukiue H, Mizuno K.et al Elevated serum epidermal growth factor receptor level is correlated with lymph node metastasis in lung cancer. Int J Clin Oncol 2003879–82. [DOI] [PubMed] [Google Scholar]
- 28.Gregorc V, Ceresoli G L, Floriani I.et al Effects of gefitinib on serum epidermal growth factor receptor and HER2 in patients with advanced non‐small cell lung cancer. Clin Cancer Res 2004106006–6012. [DOI] [PubMed] [Google Scholar]
- 29.Jacot W, Pujol J L, Boher J M.et al Serum EGF‐receptor and HER‐2 extracellular domains and prognosis of non‐small‐cell lung cancer. Br J Cancer 200491430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niemiec J, Kolodziejski L, Dyczek S.et al Prognostic significance of epidermal growth factor receptor in surgically treated squamous cell lung cancer patients. Folia Histochem Cytobiol 200442111–118. [PubMed] [Google Scholar]
- 31.Veale D, Kerr N, Gibson G J.et al The relationship of quantitative epidermal growth factor receptor expression in non‐small cell lung cancer to long term survival. Br J Cancer 199368162–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tateishi M, Ishida T, Kohdono S.et al Prognostic influence of the co‐expression of epidermal growth factor receptor and c‐erbB‐2 protein in human lung adenocarcinoma. Surg Oncol 19943109–113. [DOI] [PubMed] [Google Scholar]
- 33.Pastorino U, Andreola S, Tagliabue E.et al Immunocytochemical markers in stage I lung cancer: relevance to prognosis. J Clin Oncol 1997152858–2865. [DOI] [PubMed] [Google Scholar]
- 34.Rusch V, Klimstra D, Venkatraman E.et al Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non‐small cell lung cancer but does not predict tumor progression. Clin Cancer Res 19973515–522. [PubMed] [Google Scholar]
- 35.Fontanini G, Laurentiis M, Vignati S.et al Evaluation of epidermal growth factor‐related growth factors and receptors and of neoangiogenesis in completely resected stage I‐IIIA non‐small‐cell lung cancer: amphiregulin and microvessel count are independent prognostic indicators of survival. Clin Cancer Res 19984241–249. [PubMed] [Google Scholar]
- 36.Pfeiffer P, Nexo E, Bentzen S M.et al Enzyme‐linked immunosorbent assay of epidermal growth factor receptor in lung cancer: comparisons with immunohistochemistry, clinicopathological features and prognosis. Br J Cancer 19987896–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volm M, Rittgen W, Drings P. Prognostic value of ERBB‐1, VEGF, cyclin A, FOS, JUN and MYC in patients with squamous cell lung carcinomas. Br J Cancer 199877663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Amico T A, Massey M, Herndon J E.et al A biologic risk model for stage I lung cancer: immunohistochemical analysis of 408 patients with the use of ten molecular markers. J Thorac Cardiovasc Surg 1999117736–743. [DOI] [PubMed] [Google Scholar]
- 39.Fu X L, Zhu X Z, Shi D R.et al Study of prognostic predictors for non‐small cell lung cancer. Lung Cancer 199923143–152. [DOI] [PubMed] [Google Scholar]
- 40.Ohsaki Y, Tanno S, Fujita Y.et al Epidermal growth factor receptor expression correlates with poor prognosis in non‐small cell lung cancer patients with p53 overexpression. Oncol Rep 20007603–607. [DOI] [PubMed] [Google Scholar]
- 41.Brabender J, Danenberg K D, Metzger R.et al Epidermal growth factor receptor and HER2‐neu mRNA expression in non‐small cell lung cancer is correlated with survival. Clin Cancer Res 200171850–1855. [PubMed] [Google Scholar]
- 42.Piyathilake C J, Frost A R, Manne U.et al Differential expression of growth factors in squamous cell carcinoma and precancerous lesions of the lung. Clin Cancer Res 20028734–744. [PubMed] [Google Scholar]
- 43.Hirsch F R, Varella‐Garcia M, Bunn P A., Jret al Epidermal growth factor receptor in non‐small‐cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003213798–3807. [DOI] [PubMed] [Google Scholar]
- 44.Deeb G, Wang J, Ramnath N.et al Altered E‐cadherin and epidermal growth factor receptor expressions are associated with patient survival in lung cancer: a study utilizing high‐density tissue microarray and immunohistochemistry. Mod Pathol 200417430–439. [DOI] [PubMed] [Google Scholar]
- 45.Parra H S, Cavina R, Latteri F.et al Analysis of epidermal growth factor receptor expression as a predictive factor for response to gefitinib (‘Iressa', ZD1839) in non‐small‐cell lung cancer. Br J Cancer 200491208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selvaggi G, Novello S, Torri V.et al Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non‐small‐cell lung cancer. Ann Oncol 20041528–32. [DOI] [PubMed] [Google Scholar]
- 47.Shah L, Walter K L, Borczuk A C.et al Expression of syndecan‐1 and expression of epidermal growth factor receptor are associated with survival in patients with nonsmall cell lung carcinoma. Cancer 20041011632–1638. [DOI] [PubMed] [Google Scholar]
- 48.Swinson D E, Cox G, O'Byrne K J. Coexpression of epidermal growth factor receptor with related factors is associated with a poor prognosis in non‐small‐cell lung cancer. Br J Cancer 2004911301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parmar M K, Torri V, Stewart L. Extracting summary statistics to perform meta‐analyses of the published literature for survival endpoints. Stat Med 1998172815–2834. [DOI] [PubMed] [Google Scholar]
- 50.DerSimonian R, Laird N. Meta‐analysis in clinical trials. Controlled Clinical Trials 19867177–188. [DOI] [PubMed] [Google Scholar]
- 51.Egger M, Smith G D, Schneider M. Bias in meta‐analysis detected by a simple, graphical test. BMJ 1997315629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Begg C B, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994501088–1101. [PubMed] [Google Scholar]
- 53.Cline M J, Battifora H. Abnormalities of protooncogenes in non‐small cell lung cancer. Correlations with tumor type and clinical characteristics. Cancer 1987602669–2674. [DOI] [PubMed] [Google Scholar]
- 54.Sobol Re, Astarita R W, Hofeditz C.et al Epidermal growth factor receptor expression in human lung carcinomas defined by a monoclonal antibody. J Natl Cancer Inst 198779403–407. [PubMed] [Google Scholar]
- 55.Stewart L A, Parmar M K. Meta‐analysis of the literature or of individual patient data: is there a difference? Lancet 1993341418–422. [DOI] [PubMed] [Google Scholar]
- 56.Yusuf S, Peto R, Lewis J.et al Beta blockade during and after myocardial infarction: an overview of the randomized trials. Prog Cardiovasc Dis 198527335–371. [DOI] [PubMed] [Google Scholar]
- 57.Choma D, Daures J ‐ P, Quantin X.et al Aneuploidy and prognosis of non‐small‐cell lung cancer: a meta‐analysis of published data. Br J Cancer 20018514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitsudomi T, Hamajima N, Ogawa M.et al Prognostic significance of p53 alterations in patients with non‐small cell lung cancer: a meta‐analysis. Clin Cancer Res 200064055–4063. [PubMed] [Google Scholar]
- 59.Meert A P, Martin B, Delmotte P.et al The role of EGF‐R expression on patient survival in lung cancer: a systematic review with meta‐analysis. Eur Respir J 200220975–981. [DOI] [PubMed] [Google Scholar]
- 60.Atkins D, Reiffen K A, Tegtmeier C L.et al Immunohistochemical detection of EGFR in paraffin‐embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. J Histochem Cytochem 200452893–901. [DOI] [PubMed] [Google Scholar]
- 61.Begg C B, Berlin J A. Publication bias: A problem in interpreting medical data. J R Stat Soc A 1988151419–463. [Google Scholar]
- 62.Earleywine M. The file drawer problem in the meta‐analysis of the subjective responses to alcohol. Am J Psychiatry 19931501435–1436. [DOI] [PubMed] [Google Scholar]
- 63.Giaccone G, Herbst R S, Manegold C.et al Gefitinib in combination with gemcitabine and cisplatin in advanced non‐small‐cell lung cancer: a phase III trial ‐ INTACT 1. J Clin Oncol 200422777–784. [DOI] [PubMed] [Google Scholar]
- 64.Herbst R S, Giaccone G, Schiller J H.et al Gefitinib in combination with paclitaxel and carboplatin in advanced non‐small‐cell lung cancer: a phase III trial ‐ INTACT 2. J Clin Oncol 200422785–794. [DOI] [PubMed] [Google Scholar]
- 65.Paez J G, Janne P A, Lee J C.et al EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 20043041497–1500. [DOI] [PubMed] [Google Scholar]
- 66.Lynch T J, Bell D W, Sordella R.et al Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 20043502129–2139. [DOI] [PubMed] [Google Scholar]
- 67.Pao W, Miller V, Zakowski M.et al EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 200410113306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim Y H, Ishii G, Goto K.et al Dominant papillary subtype is a significant predictor of the response to gefitinib in adenocarcinoma of the lung. Clin Cancer Res 2004107311–7317. [DOI] [PubMed] [Google Scholar]
- 69.Perez‐Soler R, Chachoua A, Hammond L A.et al Determinants of tumor response and survival with erlotinib in patients with non‐small‐cell lung cancer. J Clin Oncol 2004223238–3247. [DOI] [PubMed] [Google Scholar]