Abstract
HLA class I molecules bind peptides derived from proteins degraded in the cytoplasm and display them for surveillance by the immune system. The recognition of HLA class I molecules by natural killer (NK) cells generally inhibits the lytic process. To investigate the role of peptides in the interaction between HLA class I molecules and NK receptors, we first had to identify representative endogenous peptides. Individual peptides bound to HLA-Cw*0304 were isolated and sequenced by tandem mass spectrometry. These peptides ranged in length from 8 to 11 residues and shared an alanine at position 2 and a C-terminal leucine. The murine transporters associated with antigen processing (TAP)-deficient cell line RMA-S was transfected with HLA-Cw*0304 to test whether HLA molecules loaded with a single peptide could deliver the inhibitory signal to NK cells expressing p58.2, which is a killer cell inhibitory receptor known to interact with HLA molecules bearing the HLA-Cw3 public epitope. We found that, in the absence of exogenous peptides, the HLA-Cw*0304 transfectants were killed at levels comparable to untransfected RMA-S cells whereas protection from lysis required both HLA-Cw*0304 heavy chain expression and an exogenously added HLA-Cw*0304-binding peptide. Importantly, not only were HLA-Cw*0304-binding peptides required for protection, but the ability of individual peptides to provide protection differed widely. These studies indicate that the ability to distinguish between subsets of peptides may be a general feature of HLA class I recognition by NK cells.
Major histocompatibility complex (MHC) class I molecules are trimeric complexes consisting of a heavy chain that is noncovalently associated with both β2-microglobulin (β2m) and a small peptide, usually 8–10 amino acids in length (1, 2). The sequence of the peptide is clearly important in cytotoxic T cell recognition of the MHC class I/peptide complex through the T cell receptor (3). Natural killer (NK) cells also bear receptors for MHC class I molecules (4–7). These receptors have been shown to recognize particular groups of class I molecules and in general transmit a negative signal to the NK cell, resulting in the protection of the MHC class I-bearing cell from NK-mediated lysis (4–7). The role of peptide in this interaction is still a subject of debate (8–12). Given the absence of genetic mechanisms for generating the diversity exhibited by the known NK receptors, it seems unlikely that these receptors have evolved to discriminate between individual peptides in a manner similar to the T cell receptor. Accordingly, in studies using murine NK cells, class I recognition by the Ly49A receptor, a member of the C-type lectin superfamily (13), was found to be peptide-dependent but not peptide-specific (9, 11). Studies of human NK cells have focused on killer cell inhibitory receptors (KIR), which are members of the Ig superfamily (14–16) and are structurally unrelated to Ly49. In the case of HLA-B*2705, some but not all endogenous peptides have been shown to protect TAP-deficient target cells expressing HLA-B27 from NK-mediated lysis, suggesting that the receptor that interacts with HLA-B27 is capable of some level of discrimination between endogenous peptides bound to HLA-B27 (8, 12). In contrast to these data, NK cells have been reported to recognize empty HLA-C molecules (10).
To clarify these issues, we have isolated and characterized by mass spectrometry a number of endogenous peptides that were bound to HLA-Cw*0304 and assessed their ability to confer protection from NK-mediated lysis. Functional studies indicated that peptides are an integral part of the HLA complex recognized by HLA-Cw3-specific NK cell clones as HLA-Cw*0304 expression alone cells does not confer protection on transporters associated with antigen processing (TAP)-deficient target cells. Not all HLA-Cw*0304 endogenous peptides conferred the same degree of resistance to lysis, and moreover, the ability of peptides to confer resistance to lysis did not correlate with the stability for particular HLA–peptide complexes.
METHODS
Antibodies.
The following mAbs were used: W6/32 (17) (anti-HLA class I), B9.12.1 (anti-HLA class I) and GL183 (anti-p58.2) (Immunotech, Westbrook, ME), 3G8 (anti-CD16) (PharMingen), Leu-19 (anti-CD56) (Becton Dickinson), UCHT1 (anti-CD3) (Dako), and fluorescein-labeled goat anti-mouse IgG F(ab′)2 (The Jackson Laboratories).
DNA Constructs.
A cDNA encoding HLA-Cw*0304 was subcloned into pRSV.neo (18) to make pRSV-HLA-Cw*0304, following PCR amplification starting from an HLA-Cw*0304 subclone in phage M13 mp18 (19) (a kind gift from Peter Parham). A bacterial expression vector (pHN1-HLA-Cw*0304) for synthesizing HLA-Cw*0304 aa 1–278 as inclusion bodies was constructed, as described previously for other HLA molecules (20). The HLA-Cw*0304 coding region of both constructs was confirmed by DNA sequencing.
Cell Lines.
pRSV-HLA-Cw*0304 was transfected into the human B cell line Hmy2.C1R (21, 22) using Lipofectin (Life Technologies, Grand Island, NY), and into the TAP-deficient murine cell line RMA-S (23) by electroporation using a Gene Pulser (Bio-Rad) set to 250 V and 960 μF. Transfectants were selected in medium containing geneticin (Life Technologies) at 1 mg/ml, and surface expression of HLA-Cw*0304 was monitored by flow cytometry using mAb W6/32. Large scale cultures of Hmy2.C1R/HLA-Cw*0304 cells were grown at the Laboratory of Cellular and Developmental Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, under the direction of J. Shiloach.
Isolation of Endogenous Peptides.
Endogenous peptides bound to HLA-Cw*0304 were isolated as described for other HLA molecules (24). Briefly, HLA complexes were purified from detergent-solubilized extracts of 1010 HLA-Cw*0304-transfected Hmy2.C1R cells by immunoaffinity chromatography using W6/32-coupled Sepharose CL-4B. After extensive washing, the HLA complexes were eluted with 0.1% trifluoroacetic acid, and the released peptides were centrifuged through a Centriprep-10 microconcentrator (Amicon) and concentrated by lyophilization to 250 μl. Peptides were separated into 100 μl fractions by reversed phase–HPLC (RP-HPLC) using a narrow-bore Vydac (Hesperia, CA) C18 column (150 × 2.1 mm, 5 μm, 330-Å pore size) with a 55 min linear gradient of 4–45% acetonitrile containing 0.1% trifluoroacetic acid and a flow rate of 250 μl/min. Individual fractions were collected, dried, and stored at −20°C prior to mass spectral analysis.
Peptide Sequence Analysis.
All mass spectrometric data were acquired on a PE-SCIEX API 300 triple-quadrupole mass spectrometer (Perkin–Elmer) equipped with a MicroIonSpray source. Dried samples were reconstituted in 15 μl of 50% acetonitrile/1% acetic acid and introduced into the ion source by infusion at 0.2 μl/min. Aliquots of 1 μl were used for mass spectrometric analysis of parent ions. Collision-activated dissociation (CAD) analysis of selected ions was used to elucidate the peptide sequence and was performed on 4 μl aliquots of sample. The program ms-tag, written by Karl Clauser and Peter Baker (available on the world wide web at http://rafael.ucsf.edu/mstag.html), was used to match CAD spectra against the protein sequence databases available at the web site. N-terminal amino acid sequence analysis was performed by automated Edman degradation as described (24). Peptides were synthesized as described (25). Purity and sequence of the synthetic peptides was established by analytical RP-HPLC analysis and by mass spectrometry.
Analysis of Peptide Binding to Class I Molecules.
An in vitro binding assay using HLA-Cw*0304 heavy chain prepared from Escherichia coli, 125I-labeled β2m (125I-β2m), and specific peptides was performed as previously described for other class I molecules (20, 25) to assess the ability of synthetic peptides to bind to HLA-Cw*0304. Briefly, inclusion bodies were purified from E. coli strain XL1-Blue (Stratagene) harboring pHN1-HLA-Cw*0304. HLA complexes were reconstituted using the HLA-Cw*0304 heavy chain (0.8 μg), 125I-β2m (10,000 cpm), and peptides (10 μg). For these experiments, 125I-β2m was prepared from β2m synthesized in E. coli. Complex formation was analyzed by HPLC gel filtration, and the stability of HLA complexes was assessed by measuring the rate of dissociation of 125I-β2m by gel filtration at various time points (25).
Analysis of Cell Surface HLA-Cw*0304 Expression.
To induce HLA-Cw*0304 expression on the cell surface, target cells (RMA-S and RMA-S-HLA-Cw*0304) were cultured for 24 h at 27°C in 5% CO2 in Opti-MEM I medium (Life Technologies) in the presence or absence of 20 μg/ml human β2m (Sigma) and the indicated peptide (100 μM). An additional aliquot of human β2m and peptide were added after 12 h to those samples that had previously received peptide. The cells were then split into two aliquots, one of which was analyzed for class I expression by flow cytometry using the mAb B9.12.1. The second aliquot was incubated for 3 h at 37°C, at which time it was analyzed for class I expression.
Generation of NK Clones.
NK clones were derived from healthy individuals and maintained as described (26). In the case of HLA-Cw3, recognition is mediated through a KIR, defined by the GL183 mAb (anti-p58.2). Therefore, we selected for clones that were p58.2+, and tested them for their ability to lyse the HLA-class I negative cell line 721.221 as well as 721.221 transfected with HLA-Cw*0304. Consistent with previous data (27), NK clones lysed 721.221 cells, whereas 721.221 HLA-Cw*0304 cells were resistant to lysis by p58.2+ NK cell clones (data not shown).
Cytotoxicity Assay.
The cytotoxic activity of NK clones was assayed in a 3 h 51Cr release assay in U-bottom 96-well plates. The assay was performed at different effector-to-target cell ratios (E/T) in triplicate wells, using 5,000 target cells per well. NK cells were washed, counted, and resuspended in Iscove’s medium containing 10% human AB serum, 200 units/ml of recombinant interleukin 2 (a kind gift from Hoffman–LaRoche) and 2 mM l-glutamine, and 1 aliquot of 100 μl was added to each well. Target cells were prepared exactly as described for the cell-surface expression assay above, except that for the last 90–120 min, the cells were labeled with sodium [51Cr]chromate. Cells were washed at room temperature and resuspended in RPMI 1640 medium containing 5% fetal calf serum and 2 mM l-glutamine, and 1 aliquot of 100 μl was added to the NK cells. The plate was centrifuged for 3 min at 900 rpm and incubated at 37°C for 3 h in the presence of 5% CO2. One hundred microliters of cell-free supernatant was harvested from each well, and radioactivity was measured in a γ-counter. Percent specific lysis was calculated as 100 × (experimental cpm − spontaneous cpm)/(total cpm − spontaneous cpm). Relative lysis was calculated as 100 × (percent specific lysis of peptide-pulsed target cells + β2m)/(percent specific lysis of target cells minus peptide + β2m).
RESULTS
Isolation and Sequencing of HLA-Cw*0304-Associated Peptides.
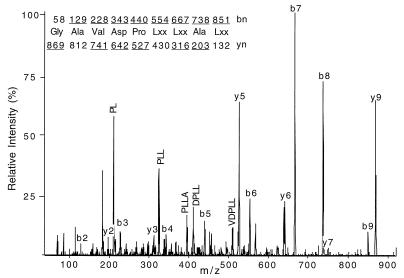
To examine the role of individual peptides bound to HLA class I molecules that protect target cells from lysis by NK cells, HLA-Cw*0304 was chosen, as the interaction between HLA-Cw3 and KIR is a paradigm for NK cell recognition of HLA molecules (27, 28). As no individual peptides bound to HLA-Cw*0304 had been identified, we isolated HLA-Cw*0304 and characterized a number of endogenously bound peptides. HLA-Cw*0304 complexes were immunoaffinity-purified from detergent lysates of Hmy2.C1R cells transfected with HLA-Cw*0304, and the associated peptides were acid-extracted and separated by RP-HPLC. The anticipated peptide-containing fractions were analyzed by electrospray ionization mass spectrometry (ESI/MS). ESI/MS analysis showed the presence of at least 40 peptides whose molecular weight fell into the mass range appropriate for 8- to 11-mer peptides. CAD analysis performed on selected ions defined plausible amino acid sequences (29). A representative CAD spectrum is shown in Fig. 1 for the peptide with mass to charge ratio (m/z) of 869 (peptide 8; Table 1). By this means, complete sequences for eight peptides were obtained and are reported in Table 1; partial sequence information was obtained on several others (data not shown). Peptides were 8–11 aa in length. When gene and protein sequence databases were searched for possible parent proteins for these peptides, six peptides matched human protein sequences. This allowed us to assign Leu or Ile, which is not possible from CAD spectra alone. In addition to the Leu at PΩ, all sequenced peptides contain Ala at P2. Furthermore, Tyr and an acidic amino acid (Asp or Glu) were observed frequently at P3 and P4, respectively. Peptides corresponding to the eight sequences reported in Table 1 were synthesized and showed both identical RP-HPLC retention times and CAD fragmentation spectra when compared with those derived from the HLA-Cw*0304-associated peptides, confirming that the deduced sequences were correct.
Figure 1.
CAD mass spectrum of peptide ions at m/z 869 (peptide 8). Predicted masses for fragment ions of types b and y (29) are shown above and below the deduced sequence, respectively. Ions observed in the spectrum are underlined. Interpretation of CAD spectra is fully explained elsewhere (29). Because Ile and Leu are of identical mass, they cannot be differentiated on the triple quadrupole instrument and are specified here as Lxx.
Table 1.
HLA-Cw*0304 endogenous peptide sequences
| Peptide† | (m/z) | Position, n
|
Protein of origin | t½‡, h | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||||
| 1 | 850.4 | V | A | Y | E | A | P | S | L | proteasome β chain (144-151) | 11.0 | |||
| 2 | 492.2 | I | A | I | I | P | S | K | K | L | 40S ribosomal protein S17 (37-45) | 2.0 | ||
| 3 | 775.4 | P | A | A | A | A | F | D | L§ | 0.5 | ||||
| 4 | 646.8 | Y | A | Y | D | G | K | D | Y | I | A | L | HLA-Cw*0304 α chain (140-150) | 49.0 |
| 5 | 638.7 | F | A | Y | D | G | K | D | Y | I | A | L | HLA-Cw*0401 α chain (140-150) | 34.0 |
| 6 | 983.2 | T | A | M | D | V | V | Y | A | L | histone H4 (82-90) | 6.3 | ||
| 7 | 934.1 | I | A | A | G | I | F | N | D | L¶ | 5.4 | |||
| 8 | 869.2 | G | A | V | D | P | L | L | A | L | importin α-1 subunit (204-212) | 9.8 | ||
| Motif | ||||||||||||||
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | PΩ | ||||||
| A | Y/A | D | L | |||||||||||
Synthetic analogs can form HLA-Cw*0304 complexes.
Peptides are listed in order of increasing RP-HPLC retention time.
Half-time of dissociation of β2m at 37°C. Data are the average of two or three separate experiments and are accurate to within 10%.
The amino acid at PΩ for peptide 3 was assumed to be Leu because all other HLA-Cw*0304 peptides whose sequence we have determined have Leu at PΩ.
Automated Edman sequencing of peptide 7 identified Ile at P1 and P5 and Leu at the C-terminal position of the peptide.
Anchor Residue Requirements for Peptide Binding to HLA-Cw*0304.
To verify that the sequences obtained represented endogenous peptides that could bind to HLA-Cw*0304, synthetic peptides corresponding to eight endogenous peptides were tested in an in vitro assay that measures the peptide-dependent incorporation of 125I-β2m into HLA complexes (25). These peptides formed complexes with HLA-Cw*0304, with stabilities that spanned two orders of magnitude (Table 1). For example, the half-time for dissociation of β2m ranged from 0.5 h for peptide 3 to almost 50 h for the two 11-mers (peptides 4 and 5).
To determine the anchor residues that are important for peptide binding to HLA-Cw*0304, synthetic single amino acid-substituted endogenous peptides, and poly-Ala-based peptides were tested for binding (Table 2). HLA-Cw*0304 complexes formed with AAYDAAAAA were about 100-fold less stable than complexes formed with AAYDAAAAL (0.2 vs. 16 h, Table 2), as expected because of the invariance of Leu at PΩ. In addition, we tested the binding of HQAISPRTL, which was reported as being a T cell epitope restricted to HLA-Cw3 (see Discussion). Note that HQAISPRTL retains the Leu at PΩ, but has Gln at P2. However, HQAISPRTL failed to bind to HLA-Cw*0304 in vitro, whereas the shorter peptide QAISPRTL, which retains the Ala at P2, forms HLA-Cw*0304 complexes with a t½ of about 1 h. Further data indicated that Tyr at P3, found in 40% of the endogenous peptides, is stabilizing, as AAYAAAAAL formed complexes slightly more stable than those formed with AAAAAAAAL (18 vs. 12 h, respectively). The contribution of an acidic aa at P4, found in 60% of the endogenous peptides, is negligible as AAADAAAAL and AAAAAAAAL formed equally stable complexes.
Table 2.
Half-time of dissociation of β2m from HLA-Cw*0304 complexes formed with various peptides
| Peptide sequence | t½†, h | Peptide sequence | t½†, h |
|---|---|---|---|
| AAYDAAAAL | 16.2 | GAVDPLLKL | 0.3 |
| AAYDAAAAA | 0.2 | GAVDPLKAL | 1.3 |
| AAAAAAAAL | 12.0 | TAMDVVYKL | 0.5 |
| AAADAAAAL | 12.0 | IAIDPSKKL | 3.0 |
| AAYAAAAAL | 18.0 | IAIIPSKAL | 37.0 |
| AAYDAAAL | 0.4 | HQAISPRTL | |
| AAYDAAAAAL | 1.7 | QAISPRTL | 0.8 |
| AAYDAAAAAAL | 1.9 | ||
| AAYDAAAAAAAL | 0.4 |
Half-time of dissociation of β2m at 37°C. Data are the average of two separate experiments and are accurate to within 10%.
To explain why complexes with the peptide IAIIPSKKL had a half-life of only 2 h, we tested a peptide in which the Lys at PΩ-1 was substituted with Ala. This resulted in a dramatic increase in stability (compare IAIIPSKKL and IAIIPSKAL, t½ of 2 and 37 h, respectively). In contrast, when the reverse substitution was made at PΩ-1 in two other endogenous peptides (namely Lys in place of Ala), the stability of the complex decreased by more than an order of magnitude (compare GAVDPLLAL and GAVDPLLKL, t½ of 9.8 and 0.3 h, respectively, and TAMDVVYAL and TAMDVVYKL, t½ of 6.3 and 0.5 h, respectively). Interestingly, the two endogenous 11-mers had the slowest dissociation rates (Table 1), even though 9-mers formed the most stable complexes when a series of poly-Ala peptides of different length was analyzed containing the anchor residues at P2 and PΩ. We thus conclude that 9 aa is the optimal length for HLA-Cw*0304-binding peptides, as has been found for most other human class I molecules (30), but something about the sequence of the endogenous 11-mers is unusually favorable for stable binding. In addition, P2 and PΩ act as typical dominant anchor residues, P3 acts as auxiliary anchor residue, and some aa at PΩ-1 are destabilizing.
Peptide/HLA-Cw*0304 Complexes Are Detectable on the Cell Surface of TAP-Deficient Cells After 3 h at 37°C.
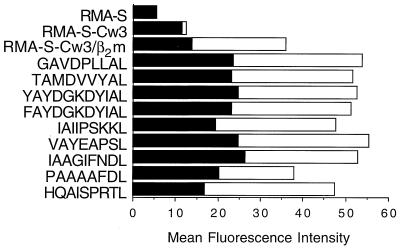
To test whether NK clones could recognize HLA-Cw*0304 molecules complexed to specific endogenous peptides, we first needed to confirm that cell surface expression of HLA-Cw*0304 complexed to specific peptides could be reconstituted under identical conditions to that used for NK cytotoxicity assays. To accomplish this we made use of the TAP-deficient cell line RMA-S transfected with HLA-Cw*0304, which does not efficiently load endogenous peptides into HLA molecules. After incubation at 27°C in the presence of β2m and a panel of synthetic peptides, the cells were washed and shifted to 37°C for 3 h in the absence of peptide and β2m. Immediately after incubation at 27°C (t = 0), HLA-Cw*0304 transfectants incubated with β2m had higher levels of cell-surface HLA-Cw*0304 as detected by flow cytometry than when β2m was absent. When certain peptides were present, the expression levels were even higher (Fig. 2). The ability of some peptides to stabilize HLA-Cw*0304 remained evident after 3 h at 37°C, although there was clearly a decline in the fluorescence signal in all cases. However, there was no significant difference in the expression levels of HLA-Cw*0304 achieved with peptides that conferred protection from NK-lysis (e.g., GAVDPLLAL) and peptides that had little protective effect (e.g., IAAGIFNDL) (see Fig. 4).
Figure 2.
Cell surface stability of HLA-Cw*0304. RMA-S cells, and RMA-S-HLA-Cw*0304 transfectants were assayed for cell surface expression at t = 0 min (open bar) and after 3 h at 37°C (filled bar). β2m alone or β2m plus the indicated peptide was added to the RMA-S-HLA-Cw*0304 transfectants as described.
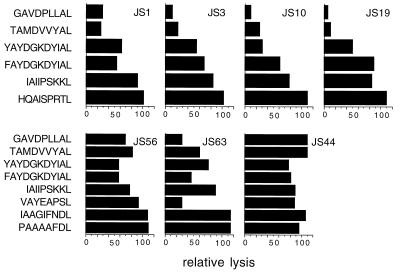
Figure 4.
A subset of endogenous peptides confers protection from lysis by p58.2+ NK clones. The same NK clones as in Fig. 2 (JS14 is not shown) were assayed in a cytotoxicity assay against RMA-S-HLA-Cw*0304 cells plus β2m and with different HLA-Cw*0304-specific peptides. The data are expressed as relative lysis (see Methods) at E/T ratio of 6:1. JS1, JS3, JS10, JS14, and JS19 NK clones were also assayed at E/T ratio 3:1 with similar results.
Requirement for Peptide in NK Cell Recognition of HLA-Cw*0304.
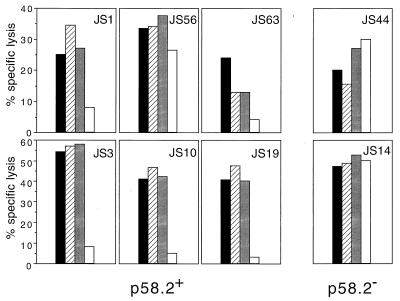
To determine whether NK cell recognition of HLA-Cw*0304 was peptide-dependent, HLA-Cw*0304 molecules were stabilized on the surface of RMA-S-HLA-Cw*0304 target cells as described above using human β2m and a synthetic peptide corresponding to the endogenous peptide, GAVDPLLAL, in medium lacking fetal calf serum. All NK clones but one lysed both RMA-S and RMA-S-HLA-Cw*0304 transfectants in the absence of added peptide even when exogenous β2m was present. (The exceptional clone, JS63, killed RMA-S, but was only about 50% as effective at killing RMA-S-HLA-Cw*0304 whether or not exogenous β2m was present.) However, coculture of RMA-S-HLA-Cw*0304 with GAVDPLLAL plus human β2m conferred significant levels of protection from NK-lysis by all but one of the p58.2+ NK clones (Fig. 3). (In the case of the exceptional NK clone, JS56, GAVDPLLAL was only marginally protective.) Finally, as might be expected, two p58.2− clones (JS14 and JS44) lysed RMA-S-HLA-Cw*0304 whether or not the peptide and β2m were present (Fig. 3). These results suggest that a bound peptide is required to protect HLA-Cw*0304+ target cells from lysis by p58.2+ NK cells.
Figure 3.
Peptides are required to induce protection from lysis by p58.2+ NK clones. p58.2+ (JS1, JS3, JS10, JS19, JS56, and JS63) and p58.2− (JS14 and JS44) NK clones were assayed in a cytotoxicity assay against RMA-S (filled bar), RMA-S-HLA-Cw*0304 (striped bar), RMA-S-HLA-Cw*0304 plus β2m (shaded bar), and RMA-S-HLA-Cw*0304 plus β2m and peptide GAVDPLLAL (open bar). The data are expressed as the percentage of specific lysis at a E/T ratio of 6:1. JS1, JS3, JS10, JS14, and JS19 NK clones were also assayed at E/T ratio 3:1 with similar results (data not shown).
Different Degrees of Peptide-Mediated Protection in the Recognition of HLA-Cw*0304 by NK Cells.
It has been previously reported that certain NK clones recognize a subset of endogenous peptides presented by HLA-B*2705 in a model system using TAP-deficient cell lines (8, 12), and that one specific KIR (NKAT3/cl11) can discriminate among different peptides bound to HLA-B*2705 (31). To test the generality of this observation, we determined whether different endogenous peptides bound to HLA-Cw*0304 would all confer resistance to NK cell-mediated lysis. We found that two peptides (GAVDPLLAL and TAMDVVYAL) induced a significant level of protection from NK cell lysis (relative lysis between 7% and 40% depending on the NK clone, except for NK clone JS56, Fig. 4). On the other hand, the other peptides were less protective, suggesting that recognition of HLA-Cw*0304 complexes by NK cells is selective and is dependent on which peptide is bound to the class I molecule. Note that NK clones JS1, JS3, JS10, and JS19 have a very similar pattern of peptide recognition to each other, while NK clone JS63 is somewhat different. For NK clone JS56, no peptide protected better than 60% relative lysis, and the 11-mer conferred the highest degree of protection. The HLA-Cw*0304-binding peptides did not prevent lysis by control p58.2− NK clones.
DISCUSSION
Although the idea that surface expression of MHC class I molecules protects cells from NK lysis is widely accepted, the role of peptides in the protection from NK-mediated lysis is still a subject of debate (8, 10, 12, 31). In this paper, we addressed whether individual peptides that bind to HLA-Cw*0304 would be equivalent in mediating inhibition of lysis by p58+ NK clones.
At the commencement of this study, the only data available about the HLA-Cw*0304 peptide-binding motif were obtained by sequencing unfractionated peptides (32).§ Consequently, it was necessary to obtain the sequences of individual representative endogenous peptides bound to HLA-Cw*0304. For this purpose, HLA-Cw*0304 complexes were isolated from Hmy2.C1R transfected cells, and the sequence of eight peptides was determined by tandem ESI/MS. We found that each peptide contained Ala at P2 and Leu at PΩ (Table 1). Binding studies carried out using differently substituted endogenous or poly-Ala peptides indicated that 9 residues is the preferred length for HLA-Cw*0304-associated peptides and confirmed that the dominant anchor residues are Ala at P2 and Leu at PΩ. The data also indicated that Tyr at P3 is an auxiliary anchor residue, while Pro at P1 and Lys at PΩ-1 are evidently unfavorable amino acids for binding to HLA-Cw*0304. Recently, three additional individual sequences of peptides eluted from HLA-Cw*0304 were reported (19). Surprisingly, none of the 11 reported individual peptide sequences has even a single instance of Pro at P3, Pro at P4, Tyr at P6, or Glu at P7, all of which had been reported to be strong anchor residues for HLA-Cw*0304 isolated from mouse cells using mAb LN29 (32). The differences between our data and previously reported data (32) may be attributable to the differences in cell type, species studied, or the mAb used in purification.
The peptide-binding motif of HLA-Cw*0304 is similar to that of HLA-Cw*0102 (34), which also has a dominant anchor of Ala at P2, presumably because these molecules share a large aa at position 9 of the class I heavy chain. Leu at PΩ has been identified as a dominant anchor residue for all five HLA-C molecules that have been studied (32, 34). Because the Hmy2.C1R cell line (HLA-A and -B negative) still expresses HLA-Cw*0401 at a low level, it could be argued that some of the peptides we isolated may have been Cw*0401-associated peptides. However, each of the peptides that we identified formed relatively stable complexes with HLA-Cw*0304. Second, our transfectants express about 15 times more HLA molecules than untransfected Hmy2.C1R cells. Finally, all known HLA-Cw*0401-associated peptides possess Tyr instead of Ala at P2 (32), presumably in part because there are smaller aa at two critical positions between the B and C pockets of the peptide-binding groove. In particular, HLA-Cw*0401 has Ser-9 and Phe-99, compared with Tyr-9 and Tyr-99 in HLA-Cw*0304.
Only one CTL epitope restricted to HLA-Cw3 has been reported (35), which had been assumed to be the nonamer, HQAISPRTL (30). This epitope had been originally localized by means of a series of truncated longer peptides (35), and no CTL studies or binding studies have been reported with HQAISPRTL itself. HQAISPRTL did not promote the binding of β2m to HLA-Cw*0304 in vitro, but the 8-mer QAISPRTL, which retains both of the dominant anchor residues, did bind to HLA-Cw*0304, suggesting that QAISPRTL is a plausible candidate for the naturally processed version of this epitope.
The role of peptides presented by MHC class I molecules in their interactions with NK cell receptors is controversial (8, 10, 12, 31). Studies with murine NK cells have shown that Ly49 molecules recognize peptide/MHC complexes, and no significant differences between peptides have been observed in their ability to confer protection from NK-mediated lysis (9, 11). However, in studies with human NK cells, pulsing NK resistant targets with specific peptides has rendered them susceptible to lysis by polyclonal NK cells suggesting that NK cells can discriminate between peptides bound by MHC class I molecules (36). Furthermore, mutations of residues of class I molecules involved directly in peptide binding have also been shown to alter susceptibility to NK cell lysis (37, 38). More recently, a single NK receptor that interacts with HLA-B27 has been shown to discriminate among endogenous peptides (31).
Our studies focus on the interaction between NK cell receptors and HLA-Cw*0304 and show that the interaction is dependent on the presence of peptide. The observed peptide dependence of NK recognition of HLA-Cw*0304 is in contrast to recent data by Mandelboim and coworkers (10). These authors concluded that NK recognition of class I does not require peptide as RMA-S cells transfected either with HLA-Cw*0602 or HLA-Cw*0702 were resistant to lysis by NK cells without the addition of exogenous peptide. There are at least four explanations for these conflicting results. First, HLA-Cw*0304 may require peptide for recognition by the p58 NK receptor, but HLA-Cw*0602 and HLA-Cw*0702 may not. Second, HLA-Cw*0602 and HLA-Cw*0702 may be appreciably loaded in a TAP-independent fashion, like HLA-A2 (39, 40). Third, there may be differences in the expression of particular NK receptors on different clones. Such considerations may also explain why the peptide recognition pattern of clone JS63 was different from the other clones. Finally, subtle differences in experimental conditions may be the explanation. We noted that RMA-S-HLA-Cw*0304 cells cultured at 27°C in the presence of 10% fetal calf serum without synthetic peptides that bind to HLA-Cw*0304 are resistant to NK-mediated lysis (data not shown). Consequently, in our studies we incubated RMA-S-HLA-Cw*0304 in serum-free medium to minimize the possibility that HLA-Cw*0304-binding peptides in the culture medium may confound the correct interpretation of the data.
Of interest, different endogenous peptides confer differing degrees of protection to RMA-S transfected with HLA-Cw*0304, indicating that NK cell receptors discriminate between different peptide/HLA-Cw*0304 complexes (Fig. 4). The ability of peptides to protect from lysis (Fig. 4) did not correlate with the stability of complexes as measured by either the in vitro assay (Table 1) or the cell surface stability assay (Fig. 2). For example, although the two 11-mer formed among the most stable complexes according to the in vitro assay and the cell surface assay, they were not as potent as GAVDPLLAL and TAMDVVYAL in conferring protection from the majority of NK clones. The observation that an NK cell can recognize a number of widely different peptides (but not all peptides) in the context of HLA-Cw*0304 supports the notion that in general NK cell recognition of class I molecules is far less specific than the recognition of class I molecules by T cells (8, 12, 31). Furthermore, the ability of particular endogenous peptides to confer protection from lysis can vary depending on the NK cell clone. Similar patterns of protection were observed among a number of NK cell clones (JS1, -3, -10, -19), while one clone (JS56) recognized a different pattern of peptides (Fig. 4). These different patterns of peptide-dependent target cell protection from NK-mediated lysis suggest that recognition of the HLA-Cw*0304/peptide complexes may involve more than one receptor. Notably, multiple loci encode slightly different p58.2 molecules and have the potential to be expressed by the same NK cell clone. In addition, the CD94/NKG2 receptors also appear to interact with HLA molecules (41, 42). Thus, the observation that the patterns of peptide recognition differ among clones (compare JS1 with JS56) could possibly reflect the contribution of different receptors and is the subject of ongoing research.
A number of studies have shown that viral infection can lead to sensitivity to NK cell-mediated cell lysis that is not simply due to a loss of class I expression (43, 44). At least two models can be used to explain these data, one of which is that viral infection induces an HLA-independent change in cell surface molecules that is recognized by NK cell receptors that induce lytic activity. The other is that viral infection results in a change in the repertoire of peptides bound to class I molecules such that they no longer are recognized by inhibitory receptors present on NK cells and hence lysis of the target cell ensues. Our studies show that p58.2+ NK cells recognize HLA Cw*0304 only if peptides are bound. Furthermore, NK cell receptors discriminate among HLA-Cw*0304 molecules when complexed with different peptides, making it a formal possibility that NK cells might sense viral infection by examination of the peptide repertoire. This differential recognition could be due to either a direct interaction with peptide, or it could be due to a peptide-induced conformational change in the HLA molecule. As this preference for particular endogenous peptides has only previously been described for HLA-B27, these studies suggest that discrimination among peptides may be a more general feature of HLA-class I recognition by NK cells.
Acknowledgments
We thank Drs. E. Fernandez, W. Magner, B. Passer, P. Posch, and J. Shuman for making useful comments on the manuscript, and A. Patamawenu for technical assistance.
ABBREVIATIONS
- β2m
β2-microglobulin
- MHC
major histocompatibility complex
- NK
natural killer
- KIR
killer cell-inhibitory receptor
- RP-HPLC
reversed phase–HPLC
- ESI/MS
electrospray ionization mass spectrometry
- CAD
collision-activated dissociation
- P1–P11
positions 1–11
- PΩ
the C-terminal amino acid of the peptide
- E/T
effector-to-target cell ratio. Standard single amino acid code is used in the text to describe peptides
- standard three-letter code is used to refer to individual amino acid.
Note Added in Proof
Since the original submission of this manuscript, similar results describing peptide selectivity in the recognition of HLA-Cw4 by p58.1 were reported by Rajagopalan and Long (45).
Footnotes
Originally, it had been believed that these peptides had been isolated from HLA-Cw*0301. Recently, it was discovered that what had been thought to be HLA-Cw*0301 was in fact HLA-Cw*0304 (33).
References
- 1.Bjorkman P J, Parham P. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 2.Madden D R. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 3.York I A, Rock K L. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Gumperz J E, Parham P. Nature (London) 1995;378:245–248. doi: 10.1038/378245a0. [DOI] [PubMed] [Google Scholar]
- 5.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari M C, Moretta L. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama W M. Proc Natl Acad Sci USA. 1995;92:3081–3085. doi: 10.1073/pnas.92.8.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottino C, Vitale M, Pende D, Biassoni R, Moretta A. Semin Immunol. 1996;7:67–73. doi: 10.1006/smim.1995.0010. [DOI] [PubMed] [Google Scholar]
- 8.Malnati M S, Peruzzi M, Parker K C, Biddison W E, Ciccone E, Moretta A, Long E O. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 9.Correa I, Raulet D H. Immunity. 1995;2:61–71. doi: 10.1016/1074-7613(95)90079-9. [DOI] [PubMed] [Google Scholar]
- 10.Mandelboim O, Reyburn H T, Valés-Gómez M, Pazmany L, Colonna M, Borsellino G, Strominger J L. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orihuela M, Margulies D H, Yokoyama W M. Proc Natl Acad Sci USA. 1996;93:11792–11797. doi: 10.1073/pnas.93.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peruzzi M, Parker K C, Long E O, Malnati M S. J Immunol. 1996;157:3350–3356. [PubMed] [Google Scholar]
- 13.Yokoyama W M, Seaman W E. Annu Rev Immunol. 1993;11:613–635. doi: 10.1146/annurev.iy.11.040193.003145. [DOI] [PubMed] [Google Scholar]
- 14.Colonna M, Samaridis J. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 15.D’Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips J H, Lanier L L. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 16.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M S, Vitale M, Bottino C, Moretta L, Moretta A, Long E O. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 17.Parham P, Barnstable C J, Bodmer W F. J Immunol. 1979;123:342–349. [PubMed] [Google Scholar]
- 18.Jacobson S, Sekaly R P, Jacobson C L, McFarland H F, Long E O. J Virol. 1989;63:1756–1762. doi: 10.1128/jvi.63.4.1756-1762.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzeng C-M, Adams E J, Gumperz J E, Percival L, Wells R S, Parham P, Barber L D. Tissue Antigens. 1996;48:325–328. doi: 10.1111/j.1399-0039.1996.tb02652.x. [DOI] [PubMed] [Google Scholar]
- 20.Parker K C, Carreno B M, Sestak L, Utz U, Biddison W E, Coligan J E. J Biol Chem. 1992;267:5451–5459. [PubMed] [Google Scholar]
- 21.Storkus W J, Howell D, Salter R D, Dawson J R, Cresswell P. J Immunol. 1987;138:1657. [PubMed] [Google Scholar]
- 22.Zemmour J, Little A-M, Schendel D J, Parham P. J Immunol. 1992;148:1941–1948. [PubMed] [Google Scholar]
- 23.Townsend A, Öhlén C, Bastin J, Ljunggren H G, Foster L, Kärre K. Nature (London) 1989;340:443–448. doi: 10.1038/340443a0. [DOI] [PubMed] [Google Scholar]
- 24.Dibrino M, Parker K C, Shiloach J, Knierman M, Lukszo J, Turner R V, Biddison W E, Coligan J E. Proc Natl Acad Sci USA. 1993;90:1508–1512. doi: 10.1073/pnas.90.4.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker K C, Dibrino M, Hull L, Coligan J E. J Immunol. 1992;149:1896–1904. [PubMed] [Google Scholar]
- 26.Brooks A G, Posch P E, Scorzelli C J, Borrego F, Coligan J E. J Exp Med. 1997;185:795–800. doi: 10.1084/jem.185.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagtmann N, Rajagopalan S, Winter C C, Peruzzi M, Long E O. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 28.Moretta A, Vitale M, Bottino C, Orengo A M, Morelli L, Augugliaro R, Barbaresi M, Ciccone E, Moretta L. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt D F, Yates J R, Shabanowitz J, Winston S, Hauer C R. Proc Natl Acad Sci USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rammensee H-G, Friede T, Stevanovic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 31.Peruzzi M, Wagtmann N, Long E O. J Exp Med. 1996;184:1585–1590. doi: 10.1084/jem.184.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falk K, Rötzschke O, Grahovac B, Schendel D, Stevanovic S, Gnau V, Jung G, Strominger J L, Rammensee H-G. Proc Natl Acad Sci USA. 1993;90:12005–12009. doi: 10.1073/pnas.90.24.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarling A L, Smith K D, Lutz C T, Lee D R. Immunogenetics. 1996;44:82–83. [PubMed] [Google Scholar]
- 34.Barber L D, Percival L, Valiante N M, Chen L, Lee C, Gumperz J E, Phillips J H, Lanier L L, Bigge J C, Parekh R B, Parham P. J Exp Med. 1996;184:735–740. doi: 10.1084/jem.184.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Littaua R A, Oldstone M B A, Takeda A, Debouck C, Wong J T, Tuazon C U, Moss B, Kievits F, Ennis F A. J Virol. 1991;65:4051–4056. doi: 10.1128/jvi.65.8.4051-4056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storkus W J, Salter R D, Cresswell P, Dawson J R. J Immunol. 1992;149:1185–1190. [PubMed] [Google Scholar]
- 37.Storkus W J, Salter R D, Alexander J, Ward F E, Ruiz R E, Cresswell P, Dawson J R. Proc Natl Acad Sci USA. 1991;88:5989–5992. doi: 10.1073/pnas.88.14.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurago Z B, Smith K D, Lutz C T. J Immunol. 1995;154:2631–2641. [PubMed] [Google Scholar]
- 39.Henderson R A, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt D F, Engelhard V H. Science. 1992;255:1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 40.Wei M L, Cresswell P. Nature (London) 1992;356:443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 41.Phillips J H, Chang C, Mattson J, Gumperz J E, Parham P, Lanier L L. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 42.Lazetic S, Chang C, Houchins J P, Lanier L L, Phillips J H. J Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 43.Kaufman D S, Schoon R A, Leibson P J. Proc Natl Acad Sci USA. 1992;89:8337–8341. doi: 10.1073/pnas.89.17.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malnati M S, Lusso P, Ciccone E, Moretta A, Moretta L, Long E O. J Exp Med. 1993;178:961–969. doi: 10.1084/jem.178.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajagopalan S, Long E O. J Exp Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]