Abstract
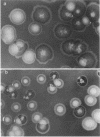
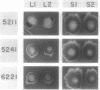
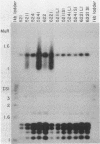
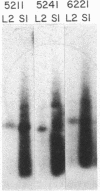
In Escherichia coli colonies, patterns of differential gene expression can be visualized by the use of Mu d(lac) fusion elements. Here we report that patterned beta-galactosidase expression in colonies of strain MS1534 resulted from a novel mechanism, spatially localized replication of the Mu dII1681 element causing lacZ transposition to active expression sites. Mu dII1681 replication did not occur constitutively with a fixed probability but was dependent on the growth history of the bacterial population. The bacteria in which Mu dII1681 replication and lacZ transposition had occurred could no longer form colonies. These results lead to several interesting conclusions about cellular differentiation during colony development and the influence of bacterial growth history on gene expression and genetic change.
Full text
PDF


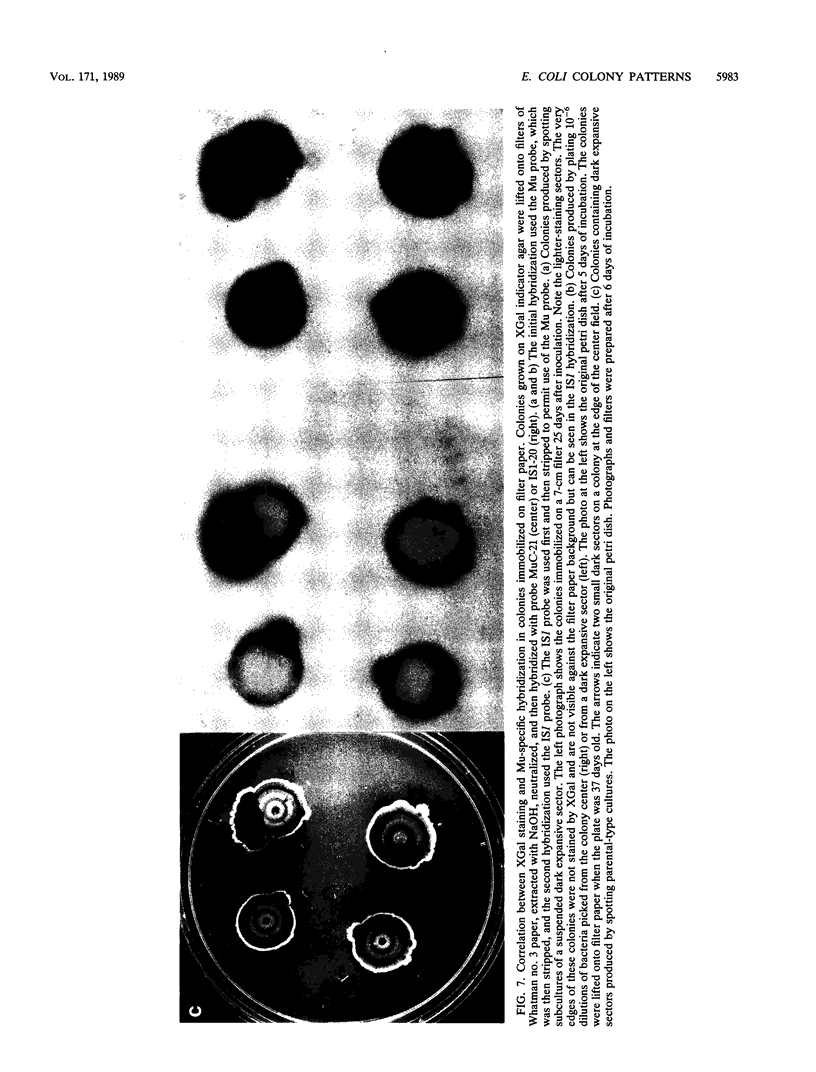



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J. M., Freitag C. S., Gander R. M., Clements J. R., Thomas V. L., Eisenstein B. I. Fimbrial phase variation and DNA rearrangements in uropathogenic isolates of Escherichia coli. Mol Biol Med. 1986 Dec;3(6):495–508. [PubMed] [Google Scholar]
- Belfort M., Wulff D. L. Genetic and biochemical investigation of the Escherichia coli mutant hfl-1 which is lysogenized at high frequency by bacteriophage lambda. J Bacteriol. 1973 Jul;115(1):299–306. doi: 10.1128/jb.115.1.299-306.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer E., Silhavy T. J., Weinstock G. M. Transposition of lambda placMu is mediated by the A protein altered at its carboxy-terminal end. Gene. 1988 Nov 15;71(1):177–186. doi: 10.1016/0378-1119(88)90089-3. [DOI] [PubMed] [Google Scholar]
- Cairns J., Overbaugh J., Miller S. The origin of mutants. Nature. 1988 Sep 8;335(6186):142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb D. I. GABAergic neurons. Sci Am. 1988 Feb;258(2):82–89. doi: 10.1038/scientificamerican0288-82. [DOI] [PubMed] [Google Scholar]
- Hall B. G. Adaptive evolution that requires multiple spontaneous mutations. I. Mutations involving an insertion sequence. Genetics. 1988 Dec;120(4):887–897. doi: 10.1093/genetics/120.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Collier D. A., Kilpatrick M. W., Krause H. M. Supercoiling and integration host factor change the DNA conformation and alter the flow of convergent transcription in phage Mu. J Biol Chem. 1989 Feb 15;264(5):3035–3042. [PubMed] [Google Scholar]
- Higgins N. P., Hillyard D. Primary structure and mapping of the hupA gene of Salmonella typhimurium. J Bacteriol. 1988 Dec;170(12):5751–5758. doi: 10.1128/jb.170.12.5751-5758.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M., Bade E. G. Molecular biology of bacteriophage mu. Science. 1975 Nov 14;190(4215):624–632. doi: 10.1126/science.1103291. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. On the mu repressor and early DNA intermediates of transposition. Cold Spring Harb Symp Quant Biol. 1984;49:827–834. doi: 10.1101/sqb.1984.049.01.093. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. State of prophage Mu DNA upon induction. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3143–3147. doi: 10.1073/pnas.74.8.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. K., Barnes W. M. Colony probing as an alternative to standard sequencing as a means of direct analysis of chromosomal DNA to determine the spectrum of single-base changes in regions of known sequence. Proc Natl Acad Sci U S A. 1986 Feb;83(4):1026–1030. doi: 10.1073/pnas.83.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki B., Rhen M., Väisänen-Rhen V., Pere A., Korhonen T. K. Organization of fimbriate cells in colonies of Escherichia coli strain 3040. J Gen Microbiol. 1985 May;131(5):1263–1266. doi: 10.1099/00221287-131-5-1263. [DOI] [PubMed] [Google Scholar]
- Pato M. L., Waggoner B. T. Cellular location of Mu DNA replicas. J Virol. 1981 Apr;38(1):249–255. doi: 10.1128/jvi.38.1.249-255.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Shore S. H., Howe M. M. Mutants of Escherichia coli defective for replicative transposition of bacteriophage Mu. J Bacteriol. 1986 Sep;167(3):905–919. doi: 10.1128/jb.167.3.905-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Brinkley P. M. Programming of DNA rearrangements involving mu prophages. Cold Spring Harb Symp Quant Biol. 1984;49:313–320. doi: 10.1101/sqb.1984.049.01.037. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., Higgins N. P. Variation of beta-galactosidase expression from Mudlac elements during the development of Escherichia coli colonies. Ann Inst Pasteur Microbiol. 1988 Jan-Feb;139(1):79–103. doi: 10.1016/0769-2609(88)90098-1. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Observations on the formation of clones containing araB-lacZ cistron fusions. Mol Gen Genet. 1984;194(1-2):79–90. doi: 10.1007/BF00383501. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Organization of developing Escherichia coli colonies viewed by scanning electron microscopy. J Bacteriol. 1987 Jan;169(1):142–156. doi: 10.1128/jb.169.1.142-156.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Scanning electron microscope study of Pseudomonas putida colonies. J Bacteriol. 1985 Dec;164(3):1171–1181. doi: 10.1128/jb.164.3.1171-1181.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. The use of Mudlac transposons as tools for vital staining to visualize clonal and non-clonal patterns of organization in bacterial growth on agar surfaces. J Gen Microbiol. 1984 May;130(5):1169–1181. doi: 10.1099/00221287-130-5-1169. [DOI] [PubMed] [Google Scholar]
- Sonti R. V., Roth J. R. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics. 1989 Sep;123(1):19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling P. F., Cannon J. G., So M. Phase and antigenic variation of pili and outer membrane protein II of Neisseria gonorrhoeae. J Infect Dis. 1986 Feb;153(2):196–201. doi: 10.1093/infdis/153.2.196. [DOI] [PubMed] [Google Scholar]
- Van Leerdam E., Karreman C., van de Putte P. Ner, a cro-like function of bacteriophage Mu. Virology. 1982 Nov;123(1):19–28. doi: 10.1016/0042-6822(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Wanner B. L. Bacterial alkaline phosphatase clonal variation in some Escherichia coli K-12 phoR mutant strains. J Bacteriol. 1986 Dec;168(3):1366–1371. doi: 10.1128/jb.168.3.1366-1371.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- van Rijn P. A., Goosen N., van de Putte P. Integration host factor of Escherichia coli regulates early- and repressor transcription of bacteriophage Mu by two different mechanisms. Nucleic Acids Res. 1988 May 25;16(10):4595–4605. doi: 10.1093/nar/16.10.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]