Abstract
The recently characterised 299-residue human XLF/Cernunnos protein plays a crucial role in DNA repair by non-homologous end joining (NHEJ) and interacts with the XRCC4–DNA Ligase IV complex. Here, we report the crystal structure of the XLF (1–233) homodimer at 2.3 Å resolution, confirming the predicted structural similarity to XRCC4. The XLF coiled-coil, however, is shorter than that of XRCC4 and undergoes an unexpected reverse in direction giving rise to a short distorted four helical bundle and a C-terminal helical structure wedged between the coiled-coil and head domain. The existence of a dimer as the major species is confirmed by size-exclusion chromatography, analytical ultracentrifugation, small-angle X-ray scattering and other biophysical methods. We show that the XLF structure is not easily compatible with a proposed XRCC4:XLF heterodimer. However, we demonstrate interactions between dimers of XLF and XRCC4 by surface plasmon resonance and analyse these in terms of surface properties, amino-acid conservation and mutations in immunodeficient patients. Our data are most consistent with head-to-head interactions in a 2:2:1 XRCC4:XLF:Ligase IV complex.
Keywords: coiled-coil, homodimer, non-homologous end-joining (NHEJ), structure, XRCC4
Introduction
DNA double-strand breaks (DSBs) are extremely cytotoxic lesions that can be generated by ionising radiation, reactive oxygen species and exposure to toxic chemicals (Khanna and Jackson, 2001; Wyman and Kanaar, 2006). Left unrepaired or incorrectly repaired, this damage can cause cell death and genome rearrangements, and these can in turn lead to cancer. Notably, DSBs also arise as intermediates during programmed genome rearrangement processes, such as site-specific V(D)J recombination that generates the antigen-binding repertoire of the mammalian adaptive immune system. Two pathways are mainly used to repair DSBs: homologous recombination that uses as the DNA repair template a homologous, undamaged DNA molecule such as the sister chromatid; and non-homologous end joining (NHEJ), a mechanism that can be used throughout the cell cycle but which is of particular importance in G1 and G0 (van Gent et al, 2001).
To date, the best characterised NHEJ factors are the Ku heterodimer (consisting of Ku70 and Ku80), the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs; Gottlieb and Jackson, 1993), the Artemis endonuclease, XRCC4 and DNA Ligase IV (Sekiguchi and Ferguson, 2006). While DNA Ligase IV, XRCC4, Ku70 and Ku80 are conserved throughout all eukaryotic species known, DNA-PKcs and Artemis are not present in simpler eukaryotes such as yeast (Critchlow and Jackson, 1998). Ku80/70 heterodimers bind to broken DNA ends to initiate the NHEJ process (Featherstone and Jackson, 1999), and DNA-PKcs serves to bridge the broken DNA ends and promote ligation by XRCC4–Ligase IV. DNA-PKcs also mediates phosphorylation of Artemis, and it is thought that this allows Artemis to cleave off the damaged bases at the broken DNA ends (Lieber et al, 1997; DeFazio et al, 2002; Ma et al, 2005; Rivera-Calzada et al, 2007). After the actions of other processing enzymes such as polynucleotide kinase and DNA polymerases, the resulting DNA ends are finally ligated by DNA Ligase IV, which is bound to XRCC4 homodimer as a cofactor (Critchlow et al, 1997; Grawunder et al, 1997). In addition to causing radio-sensitivity, inherited defects in NHEJ proteins cause severe-combined immune deficiency as a result of impaired V(D)J recombination (Schwarz et al, 2003; O'Driscoll et al, 2004; Rooney et al, 2004).
Although the above proteins complete the main functions required for NHEJ, in 2003 it became apparent that there was at least one further NHEJ factor (Dai et al, 2003). Indeed, in 2006, two groups identified a previously uncharacterised 299-amino-acid residue protein, XLF/Cernunnos (henceforth called XLF) as being essential for NHEJ in human cells (Ahnesorg et al, 2006; Buck et al, 2006). This new human NHEJ protein was named ‘XRCC4-like factor (XLF)' by one of the two groups based on an analysis with the Fugue alignment method (Shi et al, 2001) that gave 95% confidence for structural similarity between XLF and XRCC4 (Z score of 4.75), despite the low sequence identity (13.7%) between the two proteins (Ahnesorg et al, 2006). The tertiary structure of XRCC4 is a homodimer with N-terminal globular head domains and long extended α-helical coiled-coil regions (Junop et al, 2000; Sibanda et al, 2001). Notably, homotypic interactions between XLF polypeptides have been established by pull-down experiments with two differently tagged versions of the protein (Ahnesorg et al, 2006; Deshpande and Wilson, 2007). In line with there being a specific relationship between XLF and XRCC4, yeast two-hybrid results and pull-down experiments suggested the existence of a large complex containing XLF, XRCC4 and Ligase IV (Ahnesorg et al, 2006). Further biochemical investigations (Lu et al, 2007; Tsai et al, 2007) subsequently supported this contention and, furthermore, indicated that residues 1–128 of XLF bind to the head domain (residues 1–119) of XRCC4 (Deshpande and Wilson, 2007). Moreover, in the presence of Ku, XLF has been shown to enhance DNA end-joining by XRCC4–Ligase IV, and was reported to regulate DNA repair activity under conditions where base mismatches exist (Tsai et al, 2007). Notably, XLF is evolutionary and functionally conserved in diverse eukaryotes, and belongs to a superfamily of proteins that also contains the Saccharomyces cerevisiae NHEJ factors Lif1 and Nej1, which interact with one another (Callebaut et al, 2006; Hentges et al, 2006).
While the suggested structural relationship between XLF and XRCC4 has led to speculation on how XLF functions in DSB repair, so far, it has not been clear whether and to what extent XRCC4 and XLF are structurally analogous, and little is known about precisely how XLF promotes NHEJ. To address these issues, we cloned, expressed and crystallised XLF, and herein describe its tertiary structure at 2.3-Å resolution. The structure reveals both similarities to and differences from the known three-dimensional structure of XRCC4. It supports the identification of the interacting region between XLF and XRCC4 suggested by biochemical studies (Deshpande and Wilson, 2007) and provides important clues as to how XLF functions in concert with the Ligase IV-XRCC4 complex to bring about NHEJ.
Results and discussion
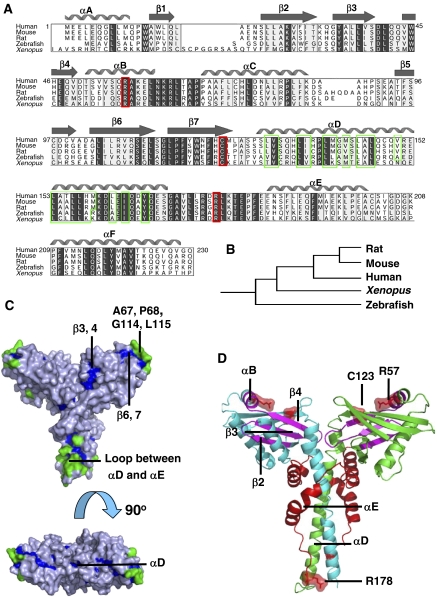
Homologues of XLF identified in human, mouse, rat, frog, fish and yeast display conserved sequence features, revealing phylogenetic relationships between the respective proteins (Figure 1A and B). Protease digestion of human full-length (299 residues) XLF revealed that it can be truncated at the C terminus to give a stable fragment of ∼27 kDa (data not shown). Results from secondary structure predictions using Jpred (Cuff et al, 1998), Coils (Lupas et al, 1991), DisPredict-EMBL (Linding et al, 2003) and Foldingdex (Prilusky et al, 2005) indicate that residues after 245 in XLF may not have a defined structure (data not shown). In view of these results, we cloned, expressed, purified and crystallised the human XLF fragment containing residues 1–233, a region that is highly conserved among all XLF orthologues (Figure 1A).
Figure 1.
Evolutionary analysis of XLF. (A) Multiple sequence alignment of XLF orthologues. Strongly conserved residues are highlighted in light grey, identical residues are in dark grey, hydrophobic residues in αD are in green frames, the reported human disease mutation sites are in red frames and secondary structure elements of human XLF are shown above the alignment. (B) Clustering of XLF orthologues generated by Evolutionary Trace Server (TraceSuite II) (Innis et al, 2000). (C) Evolutionarily conserved residues mapped onto XLF homodimer structure. Residues conserved but inaccessible to solvent are shown in blue, while those conserved and exposed to solvent are green. (D) Mapping onto XLF structure of the cancer-related mutations found in clinical cases. XLF homodimer is coloured by chain in green and cyan. Disease single-point mutation sites R57 and C123; the mutation of the polypeptide after R178 is indicated in red on both chains. Regions spanning from A25, β2 to R57, αB are magenta.
XLF wild-type crystals diffracted to 2.9-Å resolution, in space group C2, with two protomers in the asymmetric unit. Phase information was obtained with SeMet-substituted crystals by using single-wavelength anomalous diffraction (SAD). However, SeMet-substituted crystals belonged to P21 space group, with four XLF subunits in each asymmetric unit. As the SeMet-substituted crystals diffracted to a better resolution, 2.3 Å, than the wild-type crystals, the data from these crystals were used for structure determination. The R-value of the refined structure is 18.2%, and the R-free is 23.9%. The wild-type crystal structure was later solved by molecular replacement (MR) by using the model generated from SeMet-substituted structure as the template (Table I).
Table 1. Crystallographic analysis of SeMet-substituted and wild-type XLF (1–233) crystals.
| Crystal | SeMet substituted | Wild type | |
|---|---|---|---|
| X-ray diffraction data | |||
| Wavelength (Å) | 0.9807 | 0.9730 | |
| Space group | P21 | C2 | |
| Unit cell parameters | a, b, c (Å) | 63.74, 92.91, 103.69 | 111.88, 63.40, 84.90 |
| β (deg) | 106.22 | 92.71 | |
| Resolution range (Å) | High (overall) | 2.35–2.30 (50–2.30) | 2.97–2.90 (50–2.90) |
| Rsym (%) | High (overall) | 30.2 (7.9) | 50.7 (5.0) |
| Completeness (%) | High (overall) | 99.6 (99.8) | 83.4 (96.6) |
| Redundancy | High (overall) | 6.3 (7.1) | 2.5 (3.2) |
| 〈I/σ〉>3 (%) in high-resolution shell | 47.3 | 44.3 | |
| Number of reflections | 51 723 | 13 111 | |
| 〈I/σ〉 | 12.0 | 13.6 | |
| Mosaicity (deg) | 0.30 | 0.83 | |
| Wilson plot B-factor (Å2) | 43.0 | 90.0 | |
| Refinement and model quality | |||
| Resolution range (Å) | 37.01–2.30 | ||
| Number of reflections: work/test | 43 931/2000 | ||
| R-value (%) | 18.2 | ||
| R-free (%) | 23.9 | ||
| Overall mean B-factor (Å2) | 57.7 | ||
| Protein atoms | 7510 | ||
| Water and ion atoms | 235 | ||
| R.m.s.d. in bonds (Å) | 0.013 | ||
| R.m.s.d. in angles (deg) | 1.431 | ||
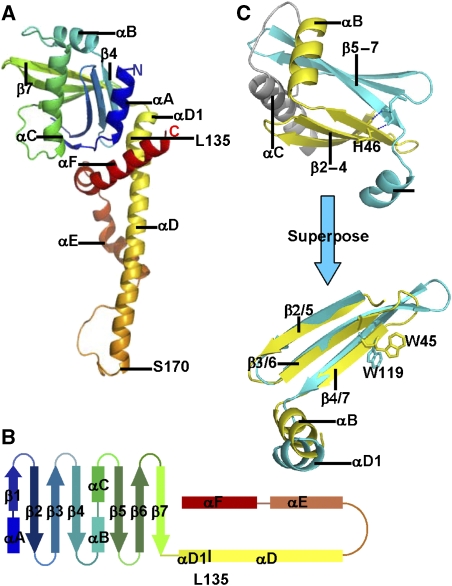
In the SeMet-substituted crystal structure, four protomers are organised as two dimers. In subunit A, residues 1–230 are clearly defined, while in subunits B, C and D residues 1–227, 1–227 and 1–229, respectively can be seen; interpretable electron density for residues 231–233 of all four subunits is absent, presumably due to disorder. Subunits A and B form a homodimer with a pseudo two-fold axis along the length of the molecule; a similar dimer is formed by subunits C and D. Each subunit has a globular head domain and a cone-shaped C-terminal part, comprised of a long α-helix, a reverse turn and two helices that wind their way around the dimeric coiled-coil (Figures 1C and 2A). Structural features plotted against the sequence alignment of XLF orthologues are shown in Figure 1A).
Figure 2.
The XLF crystal structure. (A) The structure of the XLF protomer. The secondary structure is coloured in rainbow, including an N-terminal globular head and C-terminal α-helices. The protein starts at the navy-blue α-helix and ends at the red α-helix. (B) Topology diagram of XLF protomer, secondary structure elements are in the same colour as (A). (C) Superposition of β2, β3, β4, αB (yellow) to β5, β6, β7, αD1 (cyan). The two motifs were picked up from the XLF head domain and rotated to superpose. β strands overlap well, and α-helices are in similar orientations. W45 and W119 are found at the topologically equivalent positions.
XLF has N-terminal globular head domains
The globular head of the XLF protomer (residues 1–135) contains four α-helices (αA, αB αC and αD1) and two sets of antiparallel β-sheets (β1, 2, 3, 4, and β5, 6, 7) (Figure 2A and B), organised as two β-meanders followed by helical regions: thus, the motif encompassing β2, β3, β4 and αB is similar to that containing β5, β6, β7 and αD1, and the two motifs superpose well. Remarkably, W45 of β4 and W119 of β7 are structurally equivalent and both are fully conserved across XLF orthologues (Figures 1A, C and 2C), suggesting that this structural similarity may result from an ancient gene duplication and fusion event. The two β meanders form a β-sandwich with strands lying at right angles to each other (Figure 2A). αB and αC, which are connected by a loop, lie at one end of the sandwich between the β-sheets, whilst αD1, spanning residues 128–135, forms a similar structure at the other end of the β-sandwich (Figure 2A and B). αD1 does not seem to be essential to the stability of the head domain as constructs omitting this short helix retain the ability to interact with XRCC4 (Deshpande and Wilson, 2007). The head domain resembles that of XRCC4 (Junop et al, 2000), but has not been identified elsewhere.
XLF forms a homodimer via a coiled-coil region
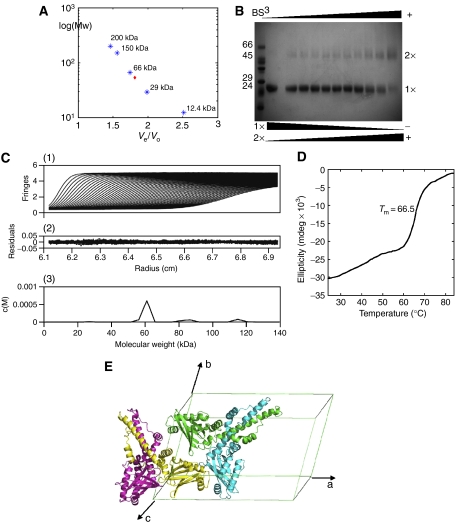
Dimerisation of XLF in solution is suggested by analytical gel-filtration chromatography and crosslinking experiments (Figure 3). By using a calibrated Superdex-200 (16/60) column, tag-free XLF (1–233) eluted at 78 ml, between the elution volumes of bovine serum albumin (66 kDa, 75 ml) and bovine carbonic anhydrase (29 kDa, 85.5 ml; Figure 3A). This indicates XLF forms a multimer, the estimated molecular weight of which is larger than that of a monomer (26.6 kDa), but smaller than that of a trimer (79.8 kDa). Further evidence of dimer formation came from bis[sulphosuccinimidyl]suberate (BS3) crosslinking experiments of XLF of the same sequence (1–233) but containing N-terminal His6 tag (Figure 3B). Two bands were found at the sizes expected for monomer and dimer, and when the mass ratio between BS3 and XLF was raised, the amount of dimer increased and monomer decreased correspondingly. Furthermore, the calculated hydrodynamic radii (12 nm), diffusion coefficient (1.98 × 10−6 m2/s) and average molecular weight (52.4 kDa) of XLF from dynamic light scattering (DLS) measurements are consistent with a protein dimer.
Figure 3.
Evidence for the XLF dimer. (A) Superdex-200 calibration curves were used to estimate the molecular weight of the XLF multimer. Proteins used for calibration are shown in blue stars, and the red diamond indicates XLF elution. (B) Crosslinking with BS3 indicated the existence of an XLF dimer, the amount of which was enhanced by increasing BS3, while the monomer decreased at the same time. (C) Sedimentation velocity profiles of XLF (1.8 mg/ml) centrifuged at 20°C and a rotor speed of 55 000 r.p.m. (1) and the residuals obtained after data fitting (2). The peak at 60 kDa (3) corresponds to the XLF dimer, which takes a relevant concentration of 92%. Data were analysed using SEDFIT program (Schuck, 2000). (D) Thermal denaturation experiment performed by CD. Tm is measured as 66.5°C. (E) XLF forms a homodimer with a two-fold axis relating protomers, and two XLF homodimers are packed in one asymmetric unit of the C2 cell.
The existence of a tightly structured XLF homodimer is confirmed by the crystal structure of both wild-type and SeMet-substituted crystal forms, which contain nearly identical homodimers. The dimer interaction interface between the two chains in the homodimer is extensive, burying ∼6100 Å2 of the molecule surface. The dimer is stabilised by interactions between the longest α-helix, αD, of each molecule through a coiled-coil structure. αD starts at P128 in all four chains, ends at S170, Y167, E169 and E169 in chains A, B, C and D, respectively and is kinked at residue L135 in each subunit (Figure 4A, left panel). The coiled-coil interface is highly hydrophobic and consists of 33 residues on each helix (Figure 1A). In each of the two dimers, the coiled-coils are stabilised by a pair of salt bridges between side chains of K160 and D161. Hydrogen bonds between residues 129–137 in αD and residues 41–43 in the loop between β3 and β4 in the head domain of the other chain also contribute to the stability of the dimer. There is high evolutionary conservation of the interface residues across different species, indicating the functional relevance of the dimeric unit and strongly suggesting that the dimeric form will persist in solution (Figure 1A and C). These extensive interactions at the protomer interface in the dimer are consistent with the independence of the far-UV circular dichroism (CD) signal of XLF concentration between 40 and 600 μg/ml and by highly cooperative thermal unfolding transition of XLF (Tm=66.5°C) (Figure 3D).
Figure 4.
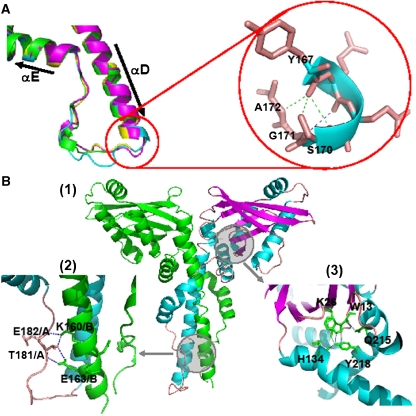
Conserved structural motifs in the C-terminal residues of XLF. (A) Loop region between αD and αE. The colour keys are set according to chains, the same as in Figure 3. Left panel: superposition of loop regions in the four chains; right panel: hydrogen bonds between the C-terminal region of αD to the following loop (Y167, Q168, S170, G171 and A172). (B) (1) Overall view of the XLF homodimer, chain A is in green, and chain B is coloured by chain in cyan (helix), magenta (sheet) and wheat (loop). Circled areas are shown in greater detail in (2) and (3). (2) Inter-chain hydrogen bonds connecting the loop of chain A and αD of chain B. (3) Hydrogen bonds between head domain residues and residues of αF.
There are intriguing interactions between the two crystallographically independent SeMet XLF dimers packed in the asymmetric unit. Thus, subunits B and D are in contact through their head domains (Figure 3E), forming three hydrogen bonds and a pair of salt bridges. The surface charge of chain B at the interface is positive, while that of chain D is negative. A similar arrangement occurs in the wild-type crystals. These observations encouraged us to investigate further whether such tetramers might exist in solution by using the more sensitive methods of sedimentation velocity and small-angle X-ray scattering (SAXS). Sedimentation velocity experiments reveal that in solution XLF is mainly (92%) a dimer as shown in Figure 3C. This is supported by SAXS intensity data, which give values of the radius of gyration (26.6±0.2 Å) and the maximum particle size (100±0.6 Å), consistent with the dimensions of an XLF dimer. The theoretical Rg value derived from the crystal structure of XLF using the program CRYSOL predicts an Rg of 26.3 Å, which is very similar to the experimental value. The theoretical Rg values for the monomer and the tetramer are 23.2 and 36.2 Å, respectively. Thus, there is no evidence that the ‘tetramer' in the crystal structure exists in solution, demonstrating that the interaction between the two head domains is weak and the tetramer is likely of crystallographic origin.
XLF C-terminal helices encircle the coiled-coil and interact with the head domain
In chain A, the coiled-coil region ends at S170 and is followed by a loop that reverses the direction of the chain towards the head domain (Figure 4A). In this structure, the Y167 carbonyl group forms hydrogen bonds with S170 and A172, while the carbonyl of Q168 contacts G171. These bonds stabilise the conformations and relative orientations of the α-helix and the loop (Figure 4A, right panel). Chains B, C and D have a similar conformation at their equivalent regions.
The loop regions following this until residue 185 differ in structure between the four protomers of the crystal asymmetric unit (Figure 4A, left panel). In chain A, the loop is a continuous random coil, while in chains B, C and D, residues 177–179 form α-helices. In addition, residues 170–173 in chain B are disordered and cannot be modelled. Hydrogen bonds, made by residues in the loop and in αD of the partner subunit, appear to guide the following helices (αE and αF) as they encircle the other molecule to form a cone-shaped homodimer (Figure 4B(1) and (2)).
αE comprises residues E186 to A201 in each chain, but a hydrogen bond between F193 and L198 gives rise to a kink in the helix allowing it to maintain its tendency to surround the coiled-coil. Two pairs of inter-chain salt bridges between K197 and E152 also help to stabilise this region. Residues following K208 in αF continue the encirclement of the coiled-coil and come close to the N terminus in the head domain. In a similar way to αE, a hydrogen bond between F210 and Q215 leads to a kink that reorients the helix. Q215 and Y218 interact with residues in the head domain to stabilise the structure through an intricate structure of three hydrogen bonds with W13, K26 and H134 (Figure 4B(1) and (3)).
Notably, all the key structural residues identified above are evolutionarily conserved in vertebrate XLF proteins, suggesting that this C-terminal structure has been selected for in evolution and is of functional significance (Figure 1A and C).
Similarities and differences between XLF and XRCC4
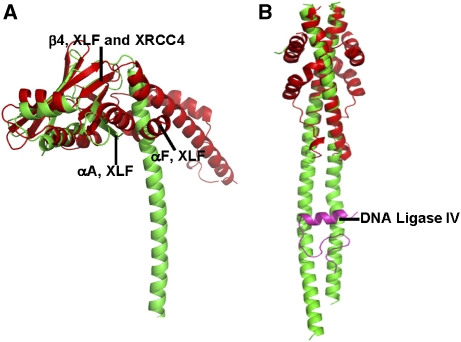
The crystal structure of the XLF homodimer is very similar to that of the XRCC4 homodimer in the head domain (Figure 5A), the main difference being that XLF has an extra α-helix (αA in Figure 2C) at its N terminus. However, the remainder of the structure differs in unexpected ways. These differences begin in the orientation of the α-helical stalks and the head domains, defined here as the angle between β4 and αD. This angle is about 130° in XLF, but it is about 85° in XRCC4 (Figure 5A). Furthermore, in the XRCC4 homodimer, the head domains interact with the stalks through van der Waals contacts and salt bridges between R3 and E125, whereas αA and αF of XLF act as wedges to position the head domains away from the α-helical stalks. Compared to the long stem-like coiled-coil region of XRCC4 (more than 120 Å), XLF has a much shorter coiled-coil of about 12 turns; and moreover, in XLF but not in XRCC4, the following sequence reverses direction to meet the N terminus. The folding of cone-shaped XLF homodimer is not similar to any known structure (Figure 5B).
Figure 5.
Superposition of XLF and XRCC4 structures. XLF red and XRCC4 green. (A) Head domains of XLF and XRCC4 superpose, especially in the antiparallel β-sheets and in the helix-turn-helix motif in the middle. XLF differs from XRCC4 in the coiled-coil region. The angle between the head domain and the coiled-coil is larger in XLF than in XRCC4 because of the insertion of αF and αA. (B) The coiled-coil in XLF is much shorter than that in XRCC4, and does not contain an equivalent region to the XRCC4–Ligase IV-binding site. DNA Ligase IV fragment bound to XRCC4 is in magenta.
The XLF complex with XRCC4–Ligase IV
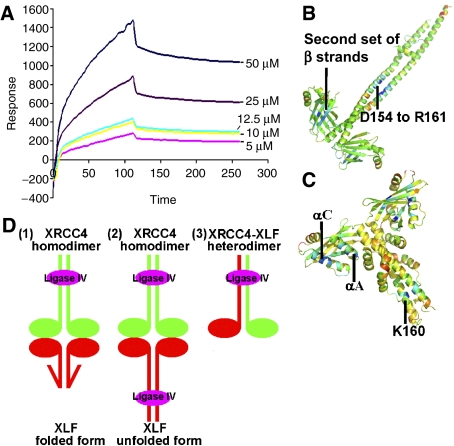
To gain insights into possible interactions between XLF and XRCC4, binding studies were performed using surface plasmon resonance on a BIAcore apparatus (BIAcore, Uppsala, Sweden). Kinetic data, evaluated using a 1:1 interaction model and obtained by exposing different concentrations of XRCC4 to XLF bound to the sensor chip (Figure 6A), showed that XLF and XRCC4 interact with an affinity of 7.8 μM.
Figure 6.
(A) XLF–XRCC4 interactions evaluated by BIAcore 2000. Sensorgrams obtained from the injections of XRCC4 over the immobilised XLF surface at concentrations of 50, 25, 12.5, 10, 5 μM. (B, C) Prediction of regions that will favour protein–protein interactions in XRCC4 and XLF structures. The darker the blue colour of a region of the dimer, the greater the probability that it acts as a binding region as indicated by ODA (Fernandez-Recio et al, 2005). XRCC4 probably interacts with other molecules through the head domains and the coiled-coil region. XLF head domains are likely to interact with other factors but also a region surrounding the conserved, K160 residue, is highlighted. (D) Possible modes of interaction between XRCC4, XLF and Ligase IV. XRCC4 molecules are shown in green, XLF is in red and Ligase IV BRCR-linker region is in magenta. (1) Linker region between Ligase IV BRCT domains binds to XRCC4's coiled-coil, folded XLF/Cernunnos contacts XRCC4 via the head domains. (2) The C termini of XLF molecules are unfolded and bind to Ligase IV in a similar way to XRCC4. Thus, there are two Ligase IV molecules in this large complex. (3) XLF and XRCC4 form a heterodimer and bind to Ligase IV in the composite coiled-coil region.
We have gained further insights into the nature of the interactions between XLF and XRCC4 by analysing the conserved surface regions among XLF and XRCC4 orthologues and by calculating the optimal docking area (ODA) (Fernandez-Recio et al, 2005). ODA predicts three potential binding regions in the XRCC4 homodimer: one spans residues D154 to R161 in the coiled-coil, while others are in the second set of β strands in each head domain (Figure 6B). ODA also predicts that XLF is likely to mediate interactions via both its head-domain and coiled-coil areas. The region surrounding the conserved K160 of XLF coiled-coil region (Figure 6C) is unlikely to be a DNA-binding region as there are also negatively charged residues in the vicinity and it is also unlikely to be a ligase-binding site, but it could be a conserved site of post-translational modification, such as ubiquitination (Figure 1A). On the other hand, the region predicted to be a binding site in the head region, especially the first and third α-helices (αA and αC) of XLF, might be involved in interacting with the XRCC4 head region, which is complementary in charge. Consistent with such a model, interactions mediated through head domains of XLF and XRCC4 have been recently indicated by yeast two-hybrid experiments (Deshpande and Wilson, 2007).
In view of the head-to-head model of XLF–XRCC4 interaction, there are two possible modes of interaction between XLF, Ligase IV and XRCC4, and these are illustrated in Figure 6D(1) and (2).
First, we must consider whether XLF could adopt a similar binding mode to Ligase IV as observed in the complex of Ligase IV with XRCC4. The XRCC4 coiled-coil includes a binding region with DNA Ligase IV spanning residues 173–195 in both chains of the XRCC4 dimer that binds with the inter-BRCT domain linker region of Ligase IV through an intricate arrangement of non-polar interactions and well-defined, often charged hydrogen bonds (Sibanda et al, 2001). Although the structure of XLF described here is well packed and identical in both SeMet-substituted and wild-type structures, there remains the possibility that this can unravel in the presence of Ligase IV and adopt a more extended coiled-coil. A radical conformational change would be consistent with the nature of the interactions between the N and C termini within one protomer chain. Whereas the residues mediating the interactions are conserved, implying a structural or functional role of the observed structure selected for in the evolution of the orthologues, the interactions involve many polar residues of the sort often observed in non-obligate complexes. As such, they would likely be fairly stable also in an unfolded form. This hypothetical model of XLF binding to Ligase IV is illustrated in Figure 6D(2).
Even if a radical conformational rearrangement of XLF were to open up a coiled-coil binding site similar to that in XRCC4, the potential binding of XLF to Ligase IV would likely be weak as there is no sequence of residues in XLF that would easily be compatible with the interactions observed between XRCC4 and Ligase IV (Sibanda et al, 2001). In view of these issues, we consider it unlikely that XLF directly interacts with the inter-BRCT domain linker region of Ligase IV. Thus, one possible model of the XLF–XRCC4–Ligase IV complex involves XLF remaining in its cone-shaped ‘folded' form and is bridged to the DNA Ligase IV linker region by XRCC4. In this scenario, the stoichiometry of XLF, XRCC4 and Ligase IV in the complex is 2:2:1 (Figure 6D(1)). We also note, however, that a variation on this model is that XLF also directly binds to a region of Ligase IV that is distinct from the Ligase IV inter-BRCT linker region. In such a model, one or two Ligase IV molecules could be associated with the XLF–XRCC4 complex.
A final potential scenario for the XLF–XRCC4–Ligase IV complex, which does not envision interactions between XLF and XRCC4 homodimers, involves the possibility that XLF forms a heterodimer with XRCC4, and that such a heterodimer mediates contacts with Ligase IV together (Figure 6D(3)). In our opinion, the hybrid coiled-coil proposed by such a structure is also not very likely to occur, because the sequence identity between the coiled-coil regions in the two proteins is very low. Furthermore, heterotypic interactions between human XLF and XRCC4 have been found to be weak and to have poor salt tolerance (Deshpande and Wilson, 2007).
Disease mutations of human XLF
Two point mutations, R57G and C123R, have been identified in immunodeficient patients with microcephaly (Buck et al, 2006). Interestingly, these residues are fully conserved among XLF homologues (Figure 1A). We have mapped these mutations onto the XLF structure (Figure 1D) and predicted their structural effects using the program SDM (Topham et al, 1997) (Table II).
Table 2. Structural effect prediction of disease mutations using SDM, negative ΔΔG refers to destabilizing mutation, while positive ΔΔG means stabilizing.
| Residues | Location | Clinical mutation | Predicted effect pseudo ΔΔG (kcal/mol) |
|---|---|---|---|
| R57 | αB | G | −3.662 |
| C123 | β7 | R | −0.651 |
| R178 | Loop between αD and αE | Deletion |
R57, which is located in the helix αB (Figure 1D), forms hydrogen bonds through its guanidinium group to the side chain of E47 (β4) and the main chain of N120 (β7). These hydrogen bonds hold the two β-meanders together, so stabilising the head domain. Loss of the arginine would be expected to destabilise the structure of the head domain and this is confirmed by a very high negative ΔΔG value predicted (Table II). In a similar way, C123 which is near the end of β7 (Figure 1D) has its side chain buried in the hydrophobic core, and this would likely be severely disrupted by the substitution of arginine, again consistent with the prediction of SDM (Table II). These observations underline the importance of the head domains to XLF function.
A mutant, R178X, in which the polypeptide chain is deleted beyond R178 in the loop region between αD and αE (Figure 1D), has also been observed in immunodeficient patients (Buck et al, 2006). This deletion must disrupt the C-terminal interactions with the head domain, giving rise to serious misfunction of XLF. Our structure indicates that a further mutant lacking A25–R57 (Buck et al, 2006) would also not be folded in the absence of residues spanning from β2 to αB (Figure 1D).
Conclusion
We have established that the XLF dimer adopts a similar overall structure to that of the XRCC4 dimer, supporting the contention that these two factors are related in function and have arisen from a common evolutionary ancestor. Nevertheless, there are important structural differences between XLF and XRCC4, suggesting strongly that the two proteins have distinct and non-overlapping functions in the NHEJ process. Our analyses also suggest that XRCC4 and XLF are unlikely to act as a heterodimer but, instead, probably associate with one another as homodimers in the complex with Ligase IV. It will now be of great interest to use the structural information at our disposal to differentiate further between the various potential models for the XLF–XRCC4–Ligase IV complex to ascertain the precise biochemical attributes of these proteins and to explore in more detail how they function in DNA repair by non-homologous end-joining.
Materials and methods
Cloning and purification
A stable XLF fragment containing the coding sequence for the TEV cleavage site and amino-acid residues 1–233 of human XLF was generated by PCR cloning into Gateway™ Destination Vectors (EMBL). The resulting plasmids included N-terminal His6-MBP tag and His6 tag, and were named as XLF441 and XLF410, respectively.
XLF441 was expressed in Escherichia coli Rosetta2 cells (Novagen). Thus, an overnight culture of 20 ml was grown at 37°C and diluted into two 1-l cultures to grow at 37°C till OD600 reached 0.6. Each culture was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 20°C overnight. Cell pellets were resuspended in 20 mM Tris pH 8.0, 300 mM NaCl, protease inhibitor (EDTA-free, Complete™; Roche). Cells were lysed by running through Emulsiflex at 2000 p.s.i. After centrifuging at 15 000 r.p.m. for 45 min, the supernatant was loaded onto 5 ml Ni-NTA beads. Imidazole (10 mM) was applied to the beads to wash away nonspecifically bound materials, and XLF was eluted with 100 mM imidazole. Eluate was dialysed in an imidazole-free buffer and treated with 400 U of TEV protease per milligram fusion protein to cleave off the N-terminal tags. Both tags in solution were removed by 5 ml Ni-NTA beads, and cleaved protein was loaded onto a Superdex-200 (16/60) column equilibrated with 20 mM Tris pH 8.0, 200 mM NaCl, 5 mM dithiothreitol (DTT). Peak fractions were concentrated to 10 mg/ml for crystallisation.
XLF410 was expressed and initially purified with Ni-NTA beads in the same way as XLF441. Its elution was loaded directly onto a Superdex-200 column without cleavage. Peak fractions were pooled and concentrated for the downstream experiments.
SeMet-substituted XLF441 was expressed by using a modified protocol. Thus, 20 ml culture was used as seed, 1 ml of these cells was diluted into 250 ml of M9 broth (containing 4.2 g/l Fe2SO4, 1 mM MgSO4, 10 ml of 40% L-glucose and 100 μl of 0.5% thiamine per litre culture as supplementary) to grow at 37°C until OD600 reached 0.3. Then L-lysine, L-threonine and L-phenylalanine (100 mg/ml each) and L-leucine, L-isoleucine, L-valine and L-selenomethionine (50 mg/ml each) were added into the cultures. Methionine synthesis was inhibited after 20 min at 37°C, and the cultures were induced with 1 mM IPTG and left shaking at 220 r.p.m. in 20°C overnight. SeMet-substituted XLF441 was purified in the same way as the native protein. After the last step, SeMet-XLF441 was concentrated to 5 mg/ml for crystallisation.
A stable XRCC4 construct containing 1–213 residues and an N-terminal His6 tag was expressed and purified using the same procedure as XLF410.
Gel filtration column calibration
A Superdex-200 (16/60) column was equilibrated with 50 mM Tris pH 7.5, 100 mM KCl; the column bed volume (Vt) was 122 ml. Gel filtration molecular weight markers (MW-GF-200; Sigma) included horse cytochrome c (12.4 kDa), bovine carbonic anhydrase (29 kDa), bovine albumin (66 kDa), yeast alcohol dehydrogenase (150 kDa) and sweet potato β-amylase (200 kDa). Column void volume (Vo) was measured with 2 mg (1 ml solution) blue dextran (2000 kDa), and the volume resulted was 43 ml. Protein samples were prepared in three groups: albumin (10 mg/ml), mixture of cytochrome c (2 mg/ml) and β-amylase (4 mg/ml), mixture of carbonic anhydrase (3 mg/ml) and alcohol dehydrogenase (5 mg/ml). Sample (1 ml) was loaded onto the column for each run. Elution volumes (Ve) of cytochrome c, carbonic anhydrase, albumin, alcohol dehydrogenase and β-amylase were 108, 85.5, 75, 67 and 63 ml, respectively.
Protein crosslinking
Purified XLF410 (1–233) was concentrated to 0.5 mg/ml and dialysed against 20 mM HEPES pH 8.0, 200 mM NaCl and 5 mM DTT. Crosslinking was performed by BS3. Stock solution containing 3% (w/v) BS3 was diluted to 1/2, 1/5, 1/10, 1/20, 1/50, 1/100, 1/200, 1/500 and 1/1000. Protein solution (10 μl) and BS3 (1 μl) solution (at different concentrations) were mixed in separate Ependorf tubes and left at room temperature for 30 min. Crosslinking was stopped by adding 1 μl of 1 M Tris pH 8.0 to each reaction and incubating for 15 min at room temperature. Protein gel loading buffer (3 ×) (6 μl) was then added and samples were loaded on 12% SDS–PAGE for analysis.
Dynamic light scattering
DLS measurements were performed using NanoS ZEN-1600 Instrument (Malvern Instruments Ltd) with 20 μM cleaved XLF441 in 20 mM Tris, 200 mM NaCl, pH 8.0. Measurements were taken at 20°C. Data were collected and analysed using the Dispersion Technology software V.5.02 (Malvern Instruments Ltd) and showed that XLF consisted of a monodisperse population of protein molecules.
Analytical ultracentrifugation
Analytical ultracentrifugation was performed on an Optima XL-I (Beckman Coulter) centrifuge with an An-60 Ti rotor, double-sector centrepieces and an interference optical system for data acquisition. Sedimentation velocity experiments were performed at a speed of 55 000 r.p.m. at 20°C. Three concentrations of isolated XLF410 were used (0.4, 0.7 and 1.8 mg/ml) and the sample volume was 400 μl. Data were analysed using SEDFIT software (Schuck, 2000). The estimations of the partial specific volumes and molecular weight were achieved by SEDINTERP software (Laue et al, 1992).
Solution X-ray scattering
High- and low-angle scattering data were collected at Station 2.1, Synchrotron Radiation Source, Daresbury Laboratory, UK, using a two-dimensional multiwire proportional counter at sample-to-detector distances of 1 and 4.25 m and an X-ray wavelength of 1.54 Å with beam currents between 120 and 200 mA. Each sample was exposed for 25 min in 30 s frames. Frames at the beginning and the end of each data collection were compared to exclude the possibility of protein aggregation and/or radiation damage. The data reduction involved radial integration, normalisation of the one-dimensional data to the intensity of the transmitted beam, correction for detector artefacts and subtraction of buffer scattering (OTOKO, SRS, Daresbury). The q-range was calibrated with an oriented specimen of wet rat-tail collagen (diffraction spacing of 670 Å) and silver behenate (diffraction spacing of 58.38 Å). XLF solutions at concentration ranging between 1 and 7 mg/ml were prepared in 20 mM Tris–HCl, 200 mM NaCl, 5 mM DTT, pH 8.0 and analysed at 4°C. The profiles collected at both camera lengths were merged so as to cover the momentum transfer interval 0.03 Å−1<q<0.77 Å−1. The modulus of the momentum transfer is defined as q=4 π sin Θ/λ, where 2Θ is the scattering angle and λ is the wavelength used. The maximum scattering angle corresponds to a nominal Bragg resolution of approximately 8 Å. The forward scattering intensity, radius of gyration Rg, the maximum particle dimension Dmax and intraparticle distance distribution function (p(r)) were calculated from the scattering data using the indirect Fourier transform method program GNOM (Svergun, 1992). The crystal structure of XLF was compared to its conformation in solution using the program CRYSOL (Svergun et al, 1995), which simulates the scattering profile from atomic coordinates and provides a goodness-of-fit relating to the experimental data by inclusion of a hydration shell.
Circular dichroism
Far-UV CD spectra were recorded on an AVIV 62-S spectropolarimeter (AVIV, NJ, USA) previously calibrated with camphorosulphonic acid and equipped with a temperature control unit. In all experiments, spectra were recorded at 20°C in a 0.1-cm quartz cell using an average time of 0.5 s, a step size of 0.5 nm, 1-nm bandwidth and averaged over 20 scans. The dependence of CD signal on protein concentration was calculated by triplicate using independent samples of concentrations ranging between 50 and 600 μg/ml. After subtraction of the buffer baseline, the CD data were normalised and reported as molar residue ellipticity. For thermal denaturation experiments, five unfolding curves were recorded upon heating from 20 to 90°C at a rate of 1°C/min, and 80 s accumulation time. The apparent melting temperature, Tm, was determined from differential melting curves of the function d[θ222](T)/dt. The concentration of protein solutions was determined from amino-acid composition analysis at the PNAC facility (Department of Biochemistry, University of Cambridge). Far-UV CD analysis of all proteins was carried out immediately after gel filtration chromatography.
Surface plasmon resonance
Biosensor surface preparation, formation and dissociation of the XLF–XRCC4 complex were monitored with a BIAcore 2000 apparatus (BIAcore AB) using HBS (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA and 0.005% surfactant P20, pH 7.4) as the running buffer. After the surface activation with a freshly prepared mixture of 50 mM N-hydroxysuccinimide and 195 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide for 4 min at 10 ml/min, purified XLF441 (cleaved) was diluted with 10 mM sodium acetate, 50 mM NaCl, pH 4.0 to a final concentration of 5 mM, and 40 μl of this sample was covalently bound to CM5 biosensor chips at 10 μl/min for 10 min; 3000 resonance units (RUs) were immobilised. Remaining activated carboxylic groups were deactivated by injecting 40 μl of 1 M ethanolamine hydrochloride, pH 8.6 for 7 min at 10 μl/min. Binding experiments were performed at 20°C in HBS at 10 μl/min (1-min injection time). After each run, the biosensor chip was regenerated using 1 M NaCl, 50 mM NaOH under the same injection condition. Five different concentrations of XRCC4 (5, 10, 12.5, 25 and 50 mM) were tested. Analysis of experimental data was performed with the interactive software BIAevaluation v3.1 (BIAcore). The simple biomolecular reaction model was used to simultaneously fit the data sets, where the analyte forms a 1:1 complex with its ligand.
Crystallisation and data collection
Crystals of XLF441 were grown using hanging-drop vapour diffusion. XLF441 (2 μl) (10 mg/ml for native protein, 5 mg/ml for SeMet-substituted protein) was mixed with same volume of well solution containing 0.1 M Bis-Tris-Propane pH 6.6, 22% PEG 6000. The volume of the well solution was 500 μl. Cryoprotectant contained 26% ethylene glycol and 74% well solution. Crystals were soaked in the cryoprotectant for a few seconds then flash frozen in liquid nitrogen.
Diffraction data of native and SeMet-substituted crystals were collected at ID29 beam line of European Synchrotron Radiation Facility. All data sets were processed by using HKL processing suite.
Structure determination and refinement
The structure was solved using SAD with SeMet-substituted crystals. Phase information was calculated by PHENIX, and 36 Se atoms were found. An initial structure was auto-built also with PHENIX, in which 60% of total amount of residues were built. The R-value was 27%, and R-free value was 31%. More residues were traced during refinements by CNS and Refmac. After six cycles of refinement and rebuilding, 903 residues and 235 water molecules were included. Because of the lack of electron density, sequence difference remains between the crystal structure and the protein sequence, as shown in Table III.
Table 3. Residues in the structure too ambiguous to identify definitively.
| Chain A | Chain B | Chain C | Chain D | |
|---|---|---|---|---|
| Left as alanine | E20 | E2, Q6, E20, K31, E169, L174 | K85, P90, E92 | E20, R81, L84, S91, E92, E185 |
| Left as glycine | P90, Q230 | D86 | H89, S91, Q227 | S170, A172, L174, D185, E182 |
| Left as serine | R176, R178 | R176, R178 | ||
| Missing | K231–Q233 | S170–T173, V228–Q233 | V228–Q233 | K85–P90, Q230–Q233 |
The coordinates of XLF have been deposited with the Protein Data Bank (PDB). The accession code is 2QM4.
Computational approaches to protein sequences and structures
Protein sequences used for alignments were obtained from the proteomics server ExPASy (Gasteiger et al, 2003). Sequences were initially aligned by ClustalW (Fukami-Kobayashi and Saito, 2002) and manually adjusted using BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Conserved and identical residues in the sequence alignments were highlighted using analysis of multiply aligned sequences (Livingstone and Barton, 1993, 1996). Secondary structure prediction was carried out using JPRED (Cuff et al, 1998) and FoldIndex (Prilusky et al, 2005). Sequences adopting coiled-coil conformation was calculated by COILS (Lupas et al, 1991). Disordered regions were predicted by DisPredict-EMBL (Linding et al, 2003). Data files of crystal structures were retrieved from the PDB (Berman et al, 2007). Phylogenetic analysis of XLF orthologues and mapping the evolutionary trace to XLF structure were done by Evolutionary Trace Server (TraceSuite II) (Innis et al, 2000). Protein surface accessibility was calculated by ODA (Fernandez-Recio et al, 2005). Superposition of protein tertiary structures are generated using COOT v1.3 (Emsley and Cowtan, 2004), and cartoon images are drawn in PyMOL v0.99rc6 (DeLano, 2002). Effect prediction of disease mutations to XLF was performed by SDM (Topham et al, 1997) with substitution tables updated by Catherine L Worth (Supplementary Figure S1).
Supplementary Material
Supplementary Figure S1
Acknowledgments
We thank Dr RN Miguel, Miss TMK Cheng and Miss CL Worth for assistance in bioinformatical analysis of the structure. We are grateful to Dr MC Moncrieffe, Dr JD Maman and Dr JG Grossmann for their kind help on analytical ultracentrifugation, surface plasmon resonance and solution X-ray scattering experiments, respectively. TL Blundell, DY Chirgadze, VM Bolanos-Garcia and BL Sibanda thank the Wellcome Trust for funding on Programme Grant RG44650 ‘The structural biology of cell signalling and regulation: multiprotein systems and the achievement of high signal-to-noise ratios' which has supported this work. Y Li thanks the Cambridge Overseas Trust and Oliver Gatty Studentship for funding the PhD study. The SP Jackson laboratory is funded by grants from Cancer Research UK and the European Union.
Competing interests statementWe declare that we have no competing financial interests in relation to the submitted work.
References
- Ahnesorg P, Smith P, Jackson SP (2006) XLF interacts with the XRCC4–DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313 [DOI] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H, Markley JL (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res 35: D301–D303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P (2006) Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP (2006) Cernunnos interacts with the XRCC4 x DNA–ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem 281: 13857–13860 [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Bowater RP, Jackson SP (1997) Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol 7: 588–598 [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23: 394–398 [DOI] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ (1998) JPred: a consensus secondary structure prediction server. Bioinformatics 14: 892–893 [DOI] [PubMed] [Google Scholar]
- Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA (2003) Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci USA 100: 2462–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G (2002) Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 21: 3192–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific [Google Scholar]
- Deshpande RA, Wilson TE (2007) Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 6: 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Featherstone C, Jackson SP (1999) Ku, a DNA repair protein with multiple cellular functions? Mutat Res 434: 3–15 [DOI] [PubMed] [Google Scholar]
- Fernandez-Recio J, Totrov M, Skorodumov C, Abagyan R (2005) Optimal docking area: a new method for predicting protein–protein interaction sites. Proteins 58: 134–143 [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Saito N (2002) [How to make good use of CLUSTALW]. Tanpakushitsu Kakusan Koso 47: 1237–1239 [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72: 131–142 [DOI] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388: 492–495 [DOI] [PubMed] [Google Scholar]
- Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, Doherty AJ (2006) Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos. J Biol Chem 281: 37517–37526 [DOI] [PubMed] [Google Scholar]
- Innis CA, Shi J, Blundell TL (2000) Evolutionary trace analysis of TGF-beta and related growth factors: implications for site-directed mutagenesis. Protein Eng 13: 839–847 [DOI] [PubMed] [Google Scholar]
- Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W (2000) Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J 19: 5962–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27: 247–254 [DOI] [PubMed] [Google Scholar]
- Laue T, Shaw BD, Ridgeway TM, Pelletier SL (1992) Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: The Royal Society of Chemistry
- Lieber MR, Grawunder U, Wu X, Yaneva M (1997) Tying loose ends: roles of Ku and DNA-dependent protein kinase in the repair of double-strand breaks. Curr Opin Genet Dev 7: 99–104 [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB (2003) Protein disorder prediction: implications for structural proteomics. Structure 11: 1453–1459 [DOI] [PubMed] [Google Scholar]
- Livingstone CD, Barton GJ (1993) Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci 9: 745–756 [DOI] [PubMed] [Google Scholar]
- Livingstone CD, Barton GJ (1996) Identification of functional residues and secondary structure from protein multiple sequence alignment. Methods Enzymol 266: 497–512 [DOI] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, Lieber MR (2007) Length-dependent binding of human XLF to DNA and stimulation of XRCC4. DNA ligase IV activity. J Biol Chem 282: 11155–11162 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Ma Y, Schwarz K, Lieber MR (2005) The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 4: 845–851 [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA (2004) An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst) 3: 1227–1235 [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL (2005) FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21: 3435–3438 [DOI] [PubMed] [Google Scholar]
- Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O (2007) Structural model of full-length human Ku70–Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep 8: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Chaudhuri J, Alt FW (2004) The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev 200: 115–131 [DOI] [PubMed] [Google Scholar]
- Schuck P (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78: 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K, Ma Y, Pannicke U, Lieber MR (2003) Human severe combined immune deficiency and DNA repair. Bioessays 25: 1061–1070 [DOI] [PubMed] [Google Scholar]
- Sekiguchi JM, Ferguson DO (2006) DNA double-strand break repair: a relentless hunt uncovers new prey. Cell 124: 260–262 [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K (2001) FUGUE: sequence–structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol 310: 243–257 [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L (2001) Crystal structure of an Xrcc4–DNA ligase IV complex. Nat Struct Biol 8: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Svergun DI (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Svergun DI, Barberato C, Koch MHJ (1995) CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28: 768–773 [Google Scholar]
- Topham CM, Srinivasan N, Blundell TL (1997) Prediction of the stability of protein mutants based on structural environment-dependent amino acid substitution and propensity tables. Protein Eng 10: 7–21 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Kim SA, Chu G (2007) Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci USA 104: 7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R (2001) Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2: 196–206 [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R (2006) DNA double-strand break repair: all's well that ends well. Annu Rev Genet 40: 363–383 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1