Abstract
Bark beetles (Coleoptera: Scolytinae) are well-recognized vectors of Ophiostoma species. Three non-native bark beetle species infest various Pinus species in South Africa, and they are known to carry at least 12 different species of ophiostomatoid fungi. Some of these fungi have not been identified to species level. The aim of this study was to determine or confirm the identities of Ophiostoma species associated with bark beetles in South Africa using comparisons of DNA sequence data. Identities of Ophiostoma ips, O. floccosum, O. pluriannulatum, O. quercus and O. stenoceras were confirmed. Ophiostoma abietinum, O. piliferum and Pesotum fragrans are recognised for the first time and the new species, O. aurorae sp. nov., is described from pine-infesting bark beetles in South Africa.
Keywords: Bark beetle, Ophiostoma, phylogeny, taxonomy
INTRODUCTION
Conifer-infesting bark beetles (Coleoptera: Scolytinae) are economically important forest insects. They include many primary pest species, which can attack healthy living trees and have caused significant economic losses to the global forestry industry (Wood & Bright 1992). In South Africa, three non-native bark beetle species, Hylastes angustatus, Hylurgus ligniperda, and Orthotomicus erosus infest various Pinus spp. (Tribe 1992). They are generally considered as secondary pests, although H. angustatus may undergo maturation feeding on healthy living seedlings causing significant losses during plantation establishment (Tribe 1992).
Bark beetles are well-known vectors of fungi, especially Ophiostoma species (Six 2003, Kirisits 2004, Harrington 2005). The ophiostomatoid fungi are a polyphyletic group of morphologically similar fungi, adapted for insect dispersal. Several ophiostomatoid fungi are important pathogens of conifers (Harrington & Cobb 1988, Wingfield et al. 1993b, Jacobs & Wingfield 2001), while many others can cause sapstain on logs and freshly cut wood (Wingfield et al. 1993b). The group includes the genera Ceratocystis Ellis & Halst., Gondwanamyces G.J. Marais & M.J. Wingf., Sphaeronaemella P. Karst. and Cornuvesica C.D. Viljoen, M.J. Wingf. & K. Jacobs and their anamorphs in the Microascales (Spatafora & Blackwell 1994, Hausner et al. 2000), and Ophiostoma Syd. & P. Syd., Grosmannia Goid. and Ceratocystiopsis H.P. Upadhyay & W.B. Kendr., with their Pesotum J.L. Crane & Schokn., Leptographium Lagerb. & Melin, Sporothrix Hektoen & C.F. Perkins and Hyalorhinocladiella H.P. Upadhyay & W.B. Kendr. anamorphs in the Ophiostomatales (Zipfel et al. 2006).
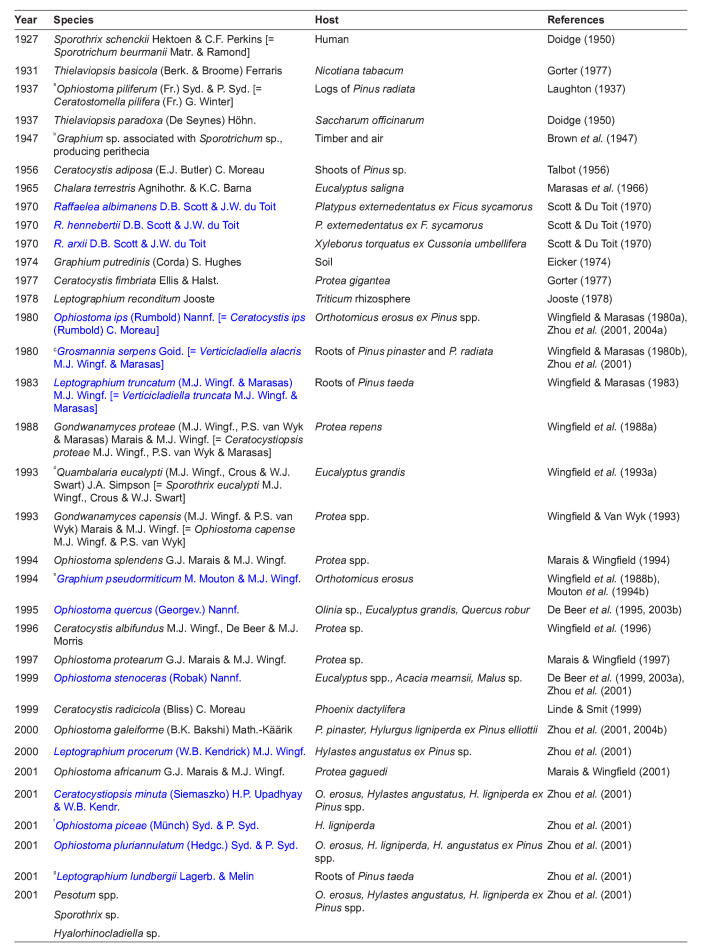
More than 30 ophiostomatoid fungi have been reported from South Africa (Table 1), of which at least 12 are associated with the three exotic pine-infesting bark beetle species in the country (Zhou et al. 2001). These fungi have been isolated from the insects or their galleries and identified based on their morphological characteristics (Zhou et al. 2001). Eight of these species belong to the genus Ophiostoma (sensu Zipfel et al. 2006) or its anamorphs. However especially those of which only the anamorphs were observed remained to be identified to species level (Zhou et al. 2001). The aim of this study was to use DNA sequence comparisons to confirm the identities of the Ophiostoma spp. (Zipfel et al. 2006) from South African pine bark beetles, previously identified based only on morphology (Zhou et al. 2001).
Table 1.
Ophiostomatoid fungi, including species with affinities to both the Microascales and Ophiostomatales, reported from South Africa. Currently accepted species names are listed first, with the name used in the original report in square brackets. Species reported as associates of bark or ambrosia beetles are printed in blue.
MATERIALS AND METHODS
Fungal isolates
Twelve isolates (Table 2) used in this study originated from a previous investigation of ophiostomatoid fungi associated with the three pine-infesting bark beetle species in South Africa (Zhou et al. 2001). All cultures are maintained in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa. A relevant sub-set of cultures has been deposited with the Centraalbureau voor Schimmelcultures (CBS), Utrecht, Netherlands.
Table 2.
Fungal isolates from pine bark beetles in South Africa used in this study.
| Species | Isolate Number |
GenBank no.
|
Host | Insect vector | Area | |
|---|---|---|---|---|---|---|
| ITS | β-tubulin | |||||
| Pesotum fragrans | aCMW 19357 | DQ396790 | Pinus patula | Hylastes angustatus | Mpumalanga | |
| Ophiostoma abietinum | CMW 397 | DQ396788 | — | Orthotomicus erosus | Western Cape | |
| O. floccosum | CMW 19358 | DQ396791 | P. elliottii | Hylurgus ligniperda | Kwazulu-Natal | |
| CMW 19359 | DQ396792 | P. patula | O. erosus | Mpumalanga | ||
| O. pluriannulatum | CMW 19360 | DQ396793 | P. elliottii | H. ligniperda | Kwazulu-Natal | |
| O. piliferum | CMW 554 | DQ396789 | — | O. erosus | Western Cape | |
| O. quercus | CMW 19361 | DQ396794 | P. patula | H. angustatus | Mpumalanga | |
| CMW 19365 | DQ396795 | P. elliottii | H. ligniperda | Kwazulu-Natal | ||
| O. stenoceras | CMW 544 | DQ396799 | — | H. angustatus | Western Cape | |
| O. aurorae | CMW 19362 | DQ396796 | DQ396800 | P. elliottii | H. angustatus | Mpumalanga |
| CMW 19363 | DQ396797 | DQ396801 | P. elliottii | H. angustatus | Mpumalanga | |
| CMW 19364 | DQ396798 | DQ396802 | P. elliottii | H. angustatus | Mpumalanga | |
Culture Collection of the Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa.
DNA sequencing and phylogenetic analyses
Single hyphal-tip cultures from the 12 isolates were grown on 2 % MEA (20 g Biolab malt extract, 20 g Biolab agar, and 1000 mL deionised water). DNA was extracted using PrepMan Ultra Sample reagent (Applied Biosystems) as described by Aghayeva et al. (2004). The ITS (internal transcribed spacer) region of the ribosomal RNA operon was amplified using primers ITS1-F (Gardes & Bruns 1993) and ITS4 (White et al. 1990). PCR products were sequenced with the same primers. Conditions for PCR amplification and sequencing reactions were as described by Zhou et al. (2004b). For comparisons, ITS sequences of closely related taxa (Table 3) were obtained from GenBank.
Table 3.
Isolates of selected species of Ophiostoma used for comparative purpose in this study.
| Species | Strain No. |
GenBank no.
|
Collector / supplier | Origin | Host / insect | |
|---|---|---|---|---|---|---|
| ITS | β-tubulin | |||||
| Leptographium guttulatum | aCMW 1310 | AY649782 | J.N. Gibbs | England | Pinus / Tomicus piniperda | |
| bCMW 742 | AY649783 | M. Morelet | France | P. sylvestris / Tomicus sp. | ||
| Pesotum fragrans | b,cCBS 279.54 | AF198248 | A. Mathiesen-Käärik | Sweden | P. sylvestris / Ips sexdentatus | |
| Ophiostoma abietinum | bCBS 125.89 | AF484453 | J.G. Marmolejo | Mexico | Abies vejari / Pseudohylesinus sp. | |
| O. dentifundum | bCBS 115790 | AY495434 | AY495445 | C. Delatour | Hungary | Quercus wood |
| CBS 115865 | AY495435 | AY495446 | T. Kowalski | Poland | Quercus robur | |
| O. floccosum | bCBS 799.73 | AF198231 | A. Käärik | Sweden | Picea or Pinus | |
| O. fusiforme | bCBS 112912 | AY280481 | AY280461 | D.N. Aghayeva | Azerbaijan | Populus nigra |
| CBS 112909 | AY280482 | AY280462 | D.N. Aghayeva | Azerbaijan | Castanea sativa | |
| O. ips | bCBS 137.36 | AY546704 | C.T. Rumbold | U.S.A. | Ips integer | |
| CMW 6418 | AY546702 | X.D. Zhou | South Africa | Pinus elliottii / Orthotomicus erosus | ||
| O. lunatum | bCBS 112927 | AY280485 | AY280466 | T. Kirisits | Austria | Carpinus betulus |
| CBS 112928 | AY280486 | AY280467 | T. Kirisits | Austria | Larix decidua | |
| O. multiannulatum | CBS 357.77 | R.W. Davidson | U.S.A. | Pinus sp. | ||
| O. narcissi | dC 1648 | AF484451 | — | U.K. | Narcissus sp. | |
| bCBS 138.50 | AY194510 | D.P. Limber | Netherlands | Narcissus sp. | ||
| O. nigrocarpum | bCBS 637.66 | AY280489 | AY280479 | R.W. Davidson | U.S.A. | Abies sp. |
| CBS 638.66 | AY280490 | AY280480 | R.W. Davidson | U.S.A. | Pseudotsuga menziesii | |
| O. piceae | bCBS 108.21 | AF198226 | E. Münch | Germany | Abies or Picea | |
| CMW 7648 | AF493249 | D.B. Redfern, J.F. Webber | U.K. | Picea sitchensis | ||
| O. piliferum | CBS 129.32 | AF221070 | H. Diddens | — | Pinus sylvestris | |
| O. pluriannulatum | CMW 75 | R.W. Davidson | U.S.A. | — | ||
| O. pulvinisporum | bCMW 9020 | AY546713 | X.D. Zhou | Mexico | P. pseudostrobus / Dendroctonus mexicanus | |
| CMW 9026 | AY546715 | X.D. Zhou | Mexico | Pinus maximinoi / Ips calligraphus | ||
| O. quercus | bCMW 2467 | AY466626 | M. Morelet | France | Quercus sp. | |
| CMW 7645 | AF493246 | T. Kirisits, E. Halmschlager | Austria | Q. robur | ||
| O. stenoceras | bCBS 237.32 | AY484462 | AY280471 | H. Robak | Norway | Pinus pulp |
| CMW 11193 | AY280493 | AY280475 | R. Farrell | New Zealand | wood | |
| Sporothrix inflata | bCBS 239.68 | AY495426 | AY495437 | W. Gams | Germany | wheatfield soil |
| CBS 841.73 | AY495431 | AY495442 | J. Grinbergs | Chile | soil | |
| S. schenckii | CMW 7612 | AY280494 | AY280476 | H.F. Vismer | South Africa | human sporotrichosis |
| CMW 7614 | AY280495 | AY280477 | H.F. Vismer | South Africa | human sporotrichosis | |
| CMW 7615 | AY280496 | AY280478 | H.F. Vismer | South Africa | human sporotrichosis | |
CMW = Culture Collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa.
Ex-type culture or authentic strain.
CBS = Culture collection of the Centraalbureau voor Schimmelcultures, Utrecht, Netherlands.
C = Culture collection of T.C. Harrington, Department of Plant Pathology, Iowa State University, U.S.A.
All sequences were aligned using MAFFT v. 5.667 (Katoh et al. 2002). Phylogenetic relationships among the isolates were determined using distance analyses in MEGA3 (http://www.megasoftware.net/). Trees were constructed using the Neighbour-joining tree-building algorithm (Saitou & Nei 1987) and rooted using GenBank sequences of Leptographium guttulatum M. J. Wingf. & K. Jacobs (AY649782 and AY649783). Bootstrap analyses (1000 replicates) were run to determine confidence levels of the branching points (Felsenstein 1985).
Three of the 12 isolates (CMW 19362, CMW 19363, and CMW 19364) grouped in a clade separate from the other isolates, all of which grouped with known taxa. For these three isolates, part of the β-tubulin gene was amplified using primers Bt2a and Bt2b (Glass & Donaldson 1995). For each of the two regions, phylogenetic analyses were done separately, followed by a distance analysis of the combined data set. A partition homogeneity test was performed in PAUP v. 4.0b8 (Phylogenetic Analyses Using Parsimony) (Swofford 2002) to determine the congruence of the two data sets.
Morphology
Isolates (CMW 19362, CMW 19363, and CMW 19364) that resided in a defined phylogenetic clade of unknown identity were grown on 2 % WA (20 g Biolab agar and 1000 mL deionised water) with sterilised pine twigs, and on 1.5 % oatmeal agar (15 g oats powder, 20 g Biolab agar and 1000 mL deionised water) to induce production of perithecia. Perithecia with ascospores were formed in two isolates (CMW 19362 and CMW 19363) on oatmeal agar. Thirty measurements were made for each structure, and the ranges and averages were computed. Anamorph structures were observed on 7-d-old slide cultures (Riddell 1950), mounted in lactophenol.
RESULTS
DNA Sequence analyses
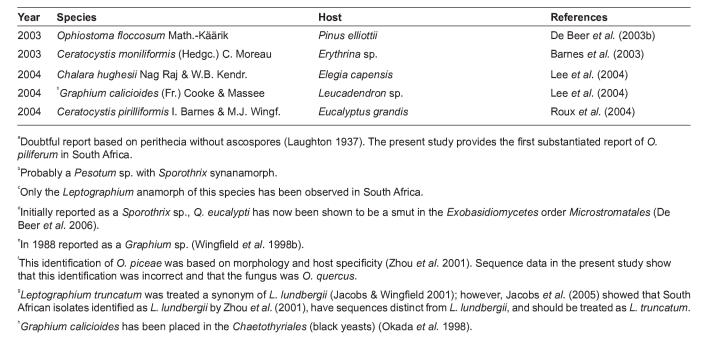
PCR of the ITS regions delivered products ranging from about 530 to 610 bp in size. Comparison of the ITS sequences with GenBank sequences confirmed the identities of seven Ophiostoma spp. (Fig. 1). These included O. stenoceras (Robak) Nannf., O. abietinum Marm. & Butin, O. piliferum (Fr.) Syd. & P. Syd., O. pluriannulatum (Hedgc.) Syd. & P. Syd., O. quercus (Georgev.) Nannf., O. floccosum Math.-Käärik, and Pesotum fragrans (Math.-Käärik) G. Okada & Seifert. The identity of O. ips (Rumbold) Nannf. (also included in the study) had previously been confirmed based on DNA sequence comparisons (Zhou et al. 2004a).
Fig. 1.
Neighbour-joining tree of Ophiostoma species associated with bark beetles in South Africa based on ITS sequences (ITS1 and ITS2 regions, as well as 5.8S rRNA gene). Isolates sequenced in this study are printed in bold. Bar = total nucleotide differences between taxa. Bootstrap values (1000 replicates) are indicated above the branches.
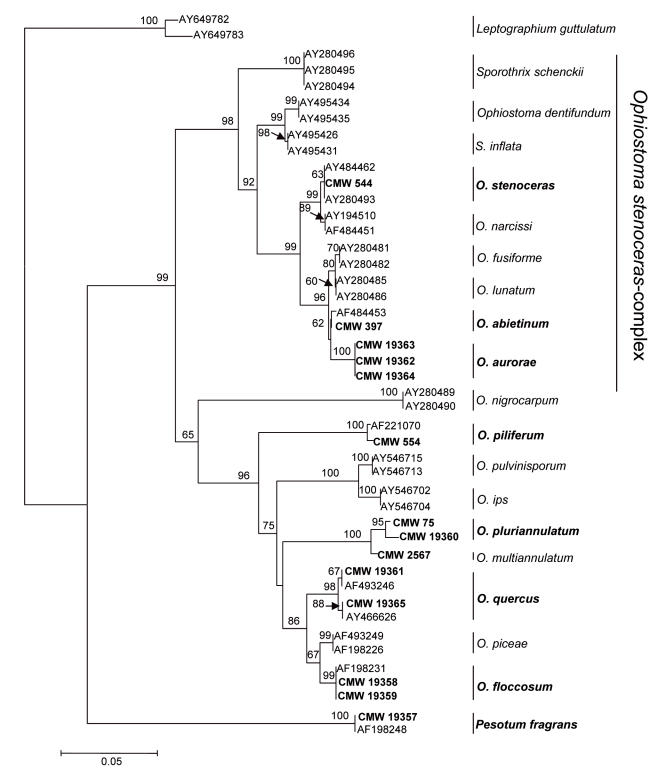
Fragments 541 bp in size from the ITS region, and 345 from the partial β-tubulin gene were amplified for the three unidentified isolates (CMW 19362, CMW 19363, and CMW 19364). The β-tubulin region included intron 5, but no intron 4 was present. This corresponds with species in the O. stenoceras -complex (Zipfel et al. 2006). Sequences of isolates representing the majority of species in this complex were thus selected for further phylogenetic analyses, with O. nigrocarpum as outgroup. Ophiostoma spp. from outside the O. stenoceras -complex were not included in these analyses because of the presence of intron 4, but no intron 5 (Zipfel et al. 2006). The partition homogeneity test (P = 0.003) confirmed that the ITS and β-tubulin data sets were congruent. Distance analyses for the combined data set showed that the three unidentified isolates grouped together with a bootstrap support of 100 % (Fig. 2), and that they either represented an undescribed species or a species for which no sequence data are available.
Fig. 2.
Neighbour-joining tree of the Ophiostoma stenoceras - Sporothrix schenckii complex of species, including O. aurorae based on the combined ITS and β-tubulin sequences. Isolates sequenced in this study are printed in bold. Bar = total nucleotide differences between taxa. Bootstrap values (1000 replicates) are indicated above the branches.
Morphology
The three isolates (CMW 19362, CMW 19363, and CMW 19364) are morphologically similar to each other and different from any other described Ophiostoma species. They produced a typical Sporothrix anamorph in culture with swollen clavate conidia. Two of the isolates produced ascomata with allantoid rounded ascospores.
TAXONOMY
Based on combined sequence comparisons of the ITS regions and partial β-tubulin gene, as well as morphology, we conclude the three isolates from H. angustatus infesting pines in South Africa represent an undescribed taxon. This is described as follows:
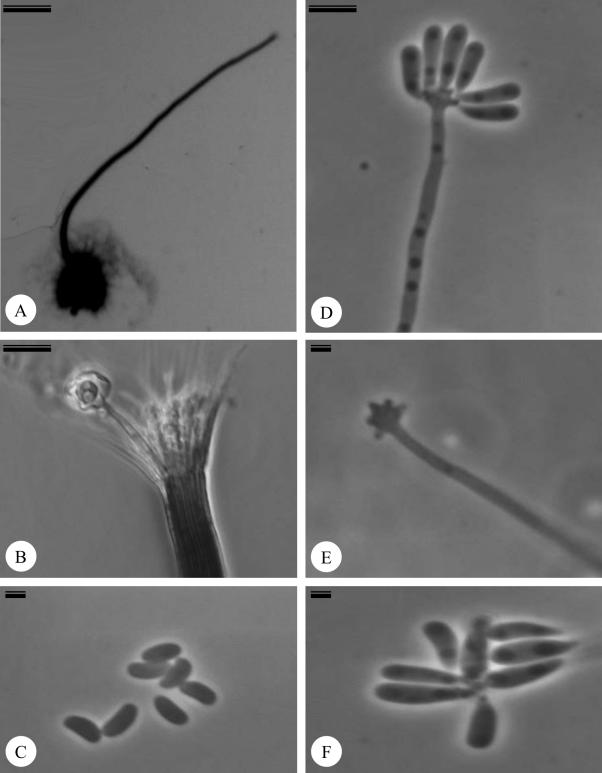
Ophiostoma aurorae X.D. Zhou & M.J. Wingf., sp. nov. MycoBank MB500888. Figs 3A–3F.
Fig. 3.
Ophiostoma aurorae (CMW 19362) on 1.5 % oatmeal agar. A. Perithecium with long neck. (Scale bar = 190 μm). B. Apex of the neck with ostiolar hyphae. (Scale bar = 15 μm). C. Allantoid round ascospores. (Scale bar = 1.5 μm). D. Conidiophore. (Scale bar = 3.5 μm). E. Conidiogenous cell. (Scale bar = 1.5 μm). F. Clavate conidia. (Scale bar = 1.5 μm).
Anamorph: Sporothrix (Fig. 3D–F).
Etymology: The type locality of this species is in Mpumalanga Province, South Africa. In siSwati, the name of the province means “the place where the sun rises”. Aurora was the Roman (Latin) goddess of dawn, so the specific epithet is an oblique reference to the type locality.
Coloniae in agaro 1.5 % avenae in medio 45 mm diam aetate duarum hebdomadum in 25 °C, laete hyalinae vel albae. Mycelium aerium adest. Ascomata superficialia vel subimmersa in agaro 1.5 % avenae. Bases perithiciorum globosae, obscurae, 130–220(–350) μm diam, hyphis laete griseis 65–150(–280) μm longis, 1.5–2.0 (–2.5) μm latis ornatae. Colla peritheciorum brunnea vel nigra, laevia, 340–800 (1415) μm longa, ad basim 35–42(–58) μm, ad apicem 12–15(–27) μm lata. Hyphae ostiolares adsunt. Ascosporae hyalinae, non septatae, allantoideae, in sectione transversali rotundae, 2–3(–3.5) x 1–1.5(–2) μm.
Cellulae conidiogenae micronematae, mononematae, hyalinae, 12–60(–85) x 1.5–2(–2.5) μm, ad apicem incrassatum denticulos acres perferentes; conidia hyalina, unicellularia, clavata vel guttuliformia, 3–4.5(–8) x 1–1.5(–2.5) μm.
Ascomata with globose bases, dark, 130–220(–350) μm diam (Fig. 3A), ornamented with light grey hyphae, 65–150(–280) μm long, 1.5–2(–2.5) μm wide. Perithecial necks brown to black, smooth, 340–800 (1415) μm long, 35–42(–58) μm wide at the base, 12–15(–27) μm at the apex (Fig. 3A, B). Ostiolar hyphae present (Fig. 3B). Ascospores hyaline, aseptate, allantoid, round in side view, 2–3(–3.5) x 1–1.5(–2) μm (Fig. 3C).
Conidiogenous cells (Fig. 3D–E), micronematous, mononematous, hyaline, 12–60(–85) x 1.5–2(–2.5) μm, sharp denticles present in the swollen apical part. Conidia (Fig. 3F) hyaline, single 1–celled, clavate to guttuliform, 3–4.5(–8) x 1–1.5(–2.5) μm.
Cultural characteristics: Colonies on 1.5 % oatmeal agar reaching on average 45 mm diam in two weeks at 25 °C. Colonies light hyaline to cotton–white. Aerial mycelium present. Perithecia produced superficially on or partially immersed in 1.5 % oatmeal agar.
Substrates: Hylastes angustatus and infested bark of Pinus patula.
Distribution: Mpumalanga Province, South Africa.
Specimens examined: South Africa, Mpumalanga Province, Hylastes angustatus, Sep. 1999, X.D. Zhou, holotype PREM 58886, culture ex-type CBS 118837 = CMW 19362; paratype PREM 58887, culture ex-paratype = CMW 19363; paratype PREM 58888, culture ex-paratype CBS 118827 = CMW 19364.
DISCUSSION
Results of this study have confirmed the identities of five Ophiostoma spp. associated with the non-native pine-infesting bark beetles H. angustatus, H. ligniperda, and O. erosus in South Africa. These fungi are O. ips, O. floccosum, O. pluriannulatum, O. quercus and O. stenoceras. In addition, O. abietinum, O. piliferum and P. fragrans are recognised for the first time from South Africa. One of the fungi associated with these bark beetles represents an undescribed taxon, for which the name O. aurorae has been provided.
The three fungal species O. abietinum, O. piliferum and P. fragrans reported from South Africa for the first time, are well-known associates of conifer timber. Ophiostoma abietinum was first described from Abies vejari attacked by a Pseudohylesinus sp. in Mexico (Marmolejo & Butin 1990), and was considered as an intermediate between O. stenoceras and O. nigrocarpum (R. Davidson) De Hoog (De Beer et al. 2003a). Ophiostoma piliferum is considered economically important to the forestry industry, and a colourless mutant of this species has been marketed as biocontrol agent against sapstaining fungi (Farrell et al. 1993). Pesotum fragrans was described from galleries of Ips sexdentatus infesting Pinus sylvestris in Sweden (Mathiesen-Käärik 1953), and the species has been reported from Australia, California, Canada, and New Zealand (Harrington et al. 2001, Jacobs et al. 2003).
Ophiostoma aurorae described in this study is morphologically similar to species in the O. stenoceras-complex (De Beer et al. 2003a, Aghayeva et al. 2004, 2005). Species in the complex have typical orange-section-shaped ascospores and Sporothrix anamorphs. Ophiostoma aurorae can be distinguished from other species in the complex by its very obviously rounded ascospores and swollen clavate conidia. Its association with the root feeding scolytid bark beetle H. angustatus also appears to be a useful characteristic that might be applied in identification. In addition to its morphologically unique nature, analyses of ITS and partial β-tubulin gene sequences confirmed that O. aurorae resides in a phylogenetic clade, distinct from all morphologically similar Ophiostoma spp. for which sequence data are available.
Results of this study emphasise that a surprisingly large number of Ophiostoma spp. are associated with the three non-native conifer-infesting bark beetles accidentally introduced into South Africa. They also highlight the fact that the introduction of what might initially appear to be a single organism (plant, insect, fungus) is often considerably more complex. It seems likely that most of the fungi treated in this study are specifically associated with the insects in their areas of origin and like their insect vectors, they are also introduced exotics.
Species such as O. quercus that have a wide distribution on many woody substrates in South Africa could have invaded the bark beetle niche. It would be interesting to understand the long-term changes in such vector/fungus relationships, as has recently been found with Tomicus piniperda (Linnaeus) and Leptographium wingfieldii M. Morelet in the United States (Jacobs et al. 2004). Clearly, the bark beetle/fungal association represents a complex and dynamic environment that deserves further study.
Acknowledgments
We thank the National Research Foundation (NRF), members of Tree Protection Co-operative Programme (TPCP) and the THRIP initiative of the Department of Trade and Industry, South Africa for financial support. We also acknowledge Sappi for a fellowship awarded to the first author and Dr Hugh F. Glen for providing the Latin diagnosis.
Taxonomic novelty: Ophiostoma aurorae X.D. Zhou & M.J. Wingf. sp. nov.
References
- Aghayeva DN, Wingfield MJ, De Beer ZW, Kirisits T (2004). Two new Ophiostoma species with Sporothrix anamorphs from Austria and Azerbaijan. Mycologia 96: 866–878. [DOI] [PubMed] [Google Scholar]
- Aghayeva DN, Wingfield MJ, Kirisits T, Wingfield BD (2005). Ophiostoma dentifundum sp. nov. from oak in Europe, characterized using molecular phylogenetic data and morphology. Mycological Research 109: 1127–1136. [DOI] [PubMed] [Google Scholar]
- Barnes I, Roux J, Wingfield BD, Dudzinski MJ, Old KM, Wingfield MJ (2003). Ceratocystis pirilliformis, a new species from Eucalyptus nitens in Australia. Mycologia 95: 865–871. [PubMed] [Google Scholar]
- Brown R, Weintroub D, Simpson MW (1947). Timber as a source of sporotrichosis infection. In: Sporotrichosis infection on Mines of the Witwatersrand. A symposium. Proceedings of the Transvaal Mine Medical Officers' Association, eds). The Transvaal Chamber of Mines, Johannesburg, South Africa: 5–33.
- De Beer ZW, Begerow D, Bauer R, Pegg GS, Crous PW, Wingfield MJ (2006). Phylogeny of Quambalariaceae fam. nov., including important Eucalyptus pathogens from South Africa and Australia. Studies in Mycology 55: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer ZW, Harrington TC, Vismer HF, Wingfield BD, Wingfield MJ (2003a). Phylogeny of the Ophiostoma stenoceras–Sporothrix schenckii complex. Mycologia 95: 434–441. [PubMed] [Google Scholar]
- De Beer ZW, Wingfield BD, Wingfield MJ (2003b). The Ophiostoma piceae-complex in the Southern Hemisphere: a phylogenetic study. Mycological Research 107: 469–476. [DOI] [PubMed] [Google Scholar]
- De Beer ZW, Wingfield MJ, Kemp GHJ (1995). First report of Ophiostoma querci in South Africa. 32nd Annual Congress of the South African Society for Plant Pathology, 23–26 January 1994, Christiana, South Africa. South African Journal of Science 91: vi. [Google Scholar]
- De Beer ZW, Witthuhn RC, Britz H, Wingfield MJ, Wingfield BD (1999). 18S rDNA sequences place Ophiostoma isolates from hardwoods in the southern hemisphere in the species Ophiostoma stenoceras. Proceedings of the 37th Annual Congress of the Southern African Society for Plant Pathology, 17–20 January 1999, Pietermaritzburg, South Africa. South African Journal of Science 95: vi. [Google Scholar]
- Doidge EM (1950). The South African fungi and lichens to the end of 1945. Bothalia 5: 1–1094. [Google Scholar]
- Eicker A (1974). The mycoflora of an alkaline soil of the open-savannah of the Transvaal. Transactions British Mycological Society 63: 281–288. [Google Scholar]
- Farrell RL, Blanchette RA, Brush TS, Hadar Y, Iverson S, Krisa K, Wendler PA, Zimmerman W (1993). Cartapip™: a biopulping product for control of pitch and resin acid problems in pulp mills. Journal of Biotechnology 30: 115–122. [Google Scholar]
- Felsenstein J (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD (1993). ITS primers with enhanced specificity for Basidiomycetes–application to the identification of mycorrhiza and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorter GMJA (1977). Index of plant pathogens and the diseases they cause in cultivated plants in South Africa. PPRI Science Bulletin 392: 177 pp. [Google Scholar]
- Harrington TC (2005). Ecology and evolution of mycophagous bark beetles and their fungal partners. In: Insect-Fungal Associations: Ecology and Evolution (Vega FE, Blackwell M, eds). Oxford University Press, New York: 1–22.
- Harrington TC, Cobb FW (1988). Leptographium root diseases on conifers. American Phytopathological Press, St. Paul, Minnesota, U.S.A.
- Harrington TC, McNew D, Steimel J, Hofstra D, Farrell R (2001). Phylogeny and taxonomy of the Ophiostoma piceae complex and the Dutch elm disease fungi. Mycologia 93: 111–136. [Google Scholar]
- Hausner G, Reid J, Klassen GR (2000). On the phylogeny of members of Ceratocystis s.s. and Ophiostoma that possess different anamorphic states, with emphasis on the anamorph genus Leptographium, based on partial ribosomal DNA sequences. Canadian Journal of Botany 78: 903–916. [Google Scholar]
- Jacobs K, Bergdahl DR, Wingfield MJ, Halk S, Seifert KA, Bright DE, Wingfield BD (2004). Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycological Research 108: 411–418. [DOI] [PubMed] [Google Scholar]
- Jacobs K, Seifert KA, Harrison KJ, Kirisits T (2003). Identity and phylogenetic relationships of ophiostomatoid fungi associated with invasive and native Tetropium species (Coleoptera: Cerambycidae) in Atlantic Canada. Canadian Journal of Botany 81: 316–329. [Google Scholar]
- Jacobs K, Solheim H, Wingfield BD, Wingfield MJ (2005). Taxonomic re-evaluation of Leptographium lundbergii based on DNA sequence comparisons and morphology. Mycological Research 109: 1149–1161. [DOI] [PubMed] [Google Scholar]
- Jacobs K, Wingfield MJ (2001). Leptographium species – tree pathogens, insect associates and agents of blue-stain. American Phytopathological Press, St. Paul, Minnesota, U.S.A.
- Jooste WJ (1978). Leptographium reconditum sp. nov. and observations on conidiogenesis in Verticicladiella. Transactions of the British Mycological Society 70: 152–155. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisits T (2004). Fungal associates of European bark beetles with special emphasis on the ophiostomatoid fungi. In: Bark and Wood boring insects in living trees in Europe, a synthesis. (Lieutier F, Day KR, Battisti A, Grégoire JC, Evans HF, eds). Kluwer Academic Press, The Netherlands: 181–235.
- Laughton EM (1937). The incidence of fungal disease on timber trees in South Africa. South African Journal of Science 33: 377–382. [Google Scholar]
- Lee S, Mel'nik V, Taylor JE, Crous PW (2004). Diversity of saprobic hyphomycetes on Proteaceae and Restionaceae from South Africa. Fungal Diversity 17: 91–114. [Google Scholar]
- Linde C, Smit WA (1999). First report of rhizosis caused by Ceratocystis radicicola on date palms in South Africa. Plant Disease 83: 880. [DOI] [PubMed] [Google Scholar]
- Marais GJ, Wingfield MJ (1994). Fungi associated with infructescences of Protea species in South Africa, including a new species of Ophiostoma. Mycological Research 98: 369–374. [Google Scholar]
- Marais GJ, Wingfield MJ (1997). Ophiostoma protearum sp. nov. associated with Protea caffra infrutescences. Canadian Journal of Botany 75: 362–367. [Google Scholar]
- Marais GJ, Wingfield MJ (2001). Ophiostoma africanum sp. nov., and a key to ophiostomatoid species from Protea infructescences. Mycological Research 105: 240–246. [Google Scholar]
- Marasas WFO, Van der Westhuizen GCA, Van Warmelo KT, Papendorf MC (1966). New and interesting records of South African fungi. Part V. Bothalia 9: 229–243. [Google Scholar]
- Marmolejo JG, Butin H (1990). New conifer-inhabiting species of Ophiostoma and Ceratocystiopsis (Ascomycetes, Microascales) from Mexico. Sydowia 42: 193–199. [Google Scholar]
- Mathiesen-Käärik A (1953). Eine Übersicht über die gewöhnlichsten mit Borkenkäfern assoziierten Bläuepilze in Schweden und einige für Schweden neue Bläuepilze. Meddelanden från Statens Skogsforskningsinstitut 43: 1–74. [Google Scholar]
- Mouton M, Wingfield MJ, Van Wyk PWJ (1994). Graphium pseudormiticum sp. nov.: a new hyphomycete with unusual conidiogenesis. Mycological Research 98: 1272–1276. [Google Scholar]
- Okada G, Seifert KA, Takematsu A, Yamaoka Y, Miyazaki S, Tubaki K (1998). A molecular phylogenetic reappraisal of the Graphium complex based on 18S rDNA sequences. Canadian Journal of Botany 76: 1495–1506. [Google Scholar]
- Riddell RW (1950). Permanent stained mycological preparations obtained by slide culture. Mycologia 42: 265–270. [Google Scholar]
- Roux J, van Wyk M, Hatting H, Wingfield MJ (2004). Ceratocystis species infecting stem wounds on Eucalyptus grandis in South Africa. Plant Pathology 53: 414–421. [Google Scholar]
- Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Scott DB, Du Toit JW (1970). Three new Raffaelea species. Transactions of the British Mycological Society 55: 181–186. [Google Scholar]
- Six DL (2003). Bark beetle–fungus symbiosis. In: Insect Symbiosis (Bourtzis K, Miller TA, eds). CRC Press, New York: 97–114.
- Spatafora JW, Blackwell M (1994). The polyphyletic origins of ophiostomatoid fungi. Mycological Research 98: 1–9. [Google Scholar]
- Swofford DL (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts.
- Talbot PHB (1956). New and interesting records of South African fungi. Part II. Bothalia 6: 489–500. [Google Scholar]
- Tribe GD (1992). Colonization sites on Pinus radiata logs of the bark beetles, Orthotomicus erosus, Hylastes angustatus and Hylurgus ligniperda (Coleoptera: Scolytidae). Journal of the Entomological Society of Southern Africa 1: 77–84. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: genes for phylogenetics (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds). Academic Press, San Diego, California, USA: 315–322.
- Wingfield MJ, Crous PW, Swart WJ (1993a). Sporothrix eucalypti (sp. nov.), a shoot and leaf pathogen of Eucalyptus in South Africa. Mycopathologia 123: 159–164. [Google Scholar]
- Wingfield MJ, De Beer C, Visser C, Wingfield BD (1996). A new Ceratocystis species defined using morphological and ribosomal DNA sequence comparisons. Systematic and Applied Microbiology 19: 191–202. [Google Scholar]
- Wingfield MJ, Marasas WFO (1980a). Ceratocystis ips associated with Orthotomicus erosus (Coleoptera: Scolytidae) on Pinus spp. in the Cape Province of South Africa. Phytophylactica 12: 65–69. [Google Scholar]
- Wingfield MJ, Marasas WFO (1980b). Verticicladiella alacris sp. nov., associated with a root disease of pines in the South Africa. Transactions of the British Mycological Society 75: 21–28. [Google Scholar]
- Wingfield MJ, Marasas WFO (1983). Some Verticicladiella species, including V. truncata sp. nov., associated with root diseases of pine in New Zealand and South Africa. Transactions of the British Mycological Society 80: 231–236. [Google Scholar]
- Wingfield MJ, Seifert KA, Webber JF (1993b). Ceratocystis and Ophiostoma. Taxonomy, Ecology and Pathogenicity. American Phytopathological Society Press, St. Paul, Minnesota, USA.
- Wingfield MJ, Van Wyk PS (1993). A new species of Ophiostoma from Protea infructescences in South Africa. Mycological Research 97: 709–716. [Google Scholar]
- Wingfield MJ, Van Wyk PS, Marasas WFO (1988a). Ceratocystiopsis proteae sp. nov., with a new anamorph genus. Mycologia 80: 23–30. [Google Scholar]
- Wingfield MJ, Van Wyk PS, Marasas WFO (1988b). A new Graphium species with unusual conidium development. Annual meeting of the South African Society for Plant Pathology. Phytophylactica 20: 103. [Google Scholar]
- Wood SL, Bright DE (1992). A catalog of Scolytidae and Platypodidae (Coleoptera), Part 2: Taxonomic index. Great Basin Naturalist Memoirs A (13): 1–6. [Google Scholar]
- Zhou XD, De Beer ZW, Cibrián D, Wingfield BD, Wingfield MJ (2004a). Characterisation of Ophiostoma species associated with pine bark beetles from Mexico, including O. pulvinisporum sp. nov. Mycological Research 108: 690–698. [DOI] [PubMed] [Google Scholar]
- Zhou XD, De Beer ZW, Harrington TC, McNew D, Kirisits T, Wingfield BD, Wingfield MJ (2004b). Epitypification of Ophiostoma galeiforme and phylogeny of species in the O. galeiforme complex. Mycologia 96: 1306–1315. [PubMed] [Google Scholar]
- Zhou XD, De Beer ZW, Wingfield BD, Wingfield MJ (2001). Ophiostomatoid fungi associated with three pine-infesting bark beetles in South Africa. Sydowia 53: 290–300. [Google Scholar]
- Zipfel RD, De Beer ZW, Jacobs K, Wingfield BD, Wingfield MJ (2006). Multi-gene phylogenies define Ceratocystiopsis and Grosmannia distinct from Ophiostoma. Studies in Mycology 55: 77–99. [DOI] [PMC free article] [PubMed] [Google Scholar]