Abstract
Colletogloeopsis zuluensis, previously known as Coniothyrium zuluense, causes a serious stem canker disease on Eucalyptus spp. grown as non-natives in many tropical and sub-tropical countries. This stem canker disease was first reported from South Africa and it has subsequently been found on various species and hybrids of Eucalyptus in other African countries as well as in countries of South America and South-East Asia. In previous studies, phylogenetic analyses based on DNA sequence data of the ITS region suggested that all material of C. zuluensis was monophyletic. However, the occurrence of the fungus in a greater number of countries, and analyses of DNA sequences with additional isolates has challenged the notion that a single species is involved with Coniothyrium canker. The aim of this study was to consider the phylogenetic relationships amongst C. zuluensis isolates from all available locations and to support these analyses with phenotypic and morphological comparisons. Individual and combined phylogenies were constructed using DNA sequences from the ITS region, exons 3 through 6 of the β-tubulin gene, the intron of the translation elongation factor 1-α gene, and a partial sequence of the mitochondrial ATPase 6 gene. Both phylogenetic data and morphological characteristics showed clearly that isolates of C. zuluensis represent at least two taxa. One of these is C. zuluensis as it was originally described from South Africa, and we provide an epitype for it. The second species occurs in Argentina and Uruguay, and is newly described as C. gauchensis. Both fungi are serious pathogens resulting in identical symptoms. Recognising them as different species has important quarantine consequences.
Keywords: Bayesian inference, Colletogloeopsis, Eucalyptus canker, multilocus gene phylogeny, Mycosphaerella
INTRODUCTION
Colletogloeopsis zuluensis (M.J. Wingf., Crous & T.A. Cout.) M.-N. Cortinas, M.J. Wingf. & Crous (Cortinas et al. 2006) causes a serious stem canker disease on Eucalyptus species. The disease was first reported in 1987 in South Africa, and the pathogen was described as a species of Coniothyrium, namely C. zuluense M.J. Wingf., Crous & T.A. Cout. (Wingfield et al. 1997). The disease spread very rapidly through the country, initially occurring only on a single Eucalyptus grandis clone, but ultimately occurring in all parts of South Africa with a sub-tropical climate, and on a wide variety of Eucalyptus species and hybrids. Substantial research has thus been undertaken to better understand the disease and to develop disease-resistant planting stock through breeding and selection programmes (Van Zyl et al. 1997, 2002a).
Symptoms of Colletogloepsis canker are very obvious, at least at the onset of disease. Initial infections include small, circular necrotic lesions on the green stem tissue in the upper parts of trees. These lesions expand, becoming elliptical, and the dead bark covering them typically cracks, giving a “cat-eye” appearance (Fig. 1). Lesions coalesce to form large cankers that girdle the stems, giving rise to the production of epicormic shoots and ultimately trees with malformed or dead tops. Infections occur annually on the new green tissue and they penetrate the cambium to form black kino-filled pockets. Thus kino pockets with irregular borders of infected tissue can be seen within the infected wood of trees coincident with the annual rings (Fig. 1). Small black pycnidia can be seen on the surface of dead bark tissue (Fig. 1), from where black conidial tendrils exude under moist conditions. Conidia are small, aseptate and dematiaceous, appearing black in colour when seen in mass on the host or agar media.
Fig. 1.
External symptoms of the stem canker disease on E. grandis in Uruguay caused by C. gauchensis. A, B. Mature clones showing the typical lesions on the surface of the trunk. C. Distinctive black circular lesions on green twigs. D. Stem with typical cracked lesions. E. Stem showing internal symptoms below the bark lesions. F. Kino-pockets of infected tissue within the wood. G. Pycnidia on cracked lesions.
Subsequent to the discovery of Coniothyrium canker in South Africa, the disease has been found in many other countries. Its first discovery outside South Africa was in Thailand where it is associated with typical symptoms on E. camaldulensis (Van Zyl et al. 2002b). More recently, the disease has been found in other countries in Africa (Alemu et al. 2003, 2005), South and Central America (Roux et al. 2002, Alemu et al. 2004), as well as South-East Asia (Old et al. 2003, Cortinas et al. 2004, 2006) (Fig. 2). Interestingly, the disease remains unknown in the areas of origin of Eucalyptus, although it might occur there at very low and undetectable levels (Wingfield 2003, Slippers et al. 2005).
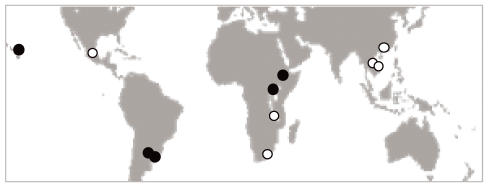
Fig. 2.
Geographical range of the collection of isolates used in this study. The map includes isolates from South Africa, Malawi, Vietnam, Thailand and China, indicated with white dots (Group 1) and isolates from Uruguay, Argentina, Hawaii-U.S.A., Ethiopia and Uganda, indicated with black dots (Group 2).
The first taxonomic treatment of C. zuluensis was based on morphological characteristics of the pathogen. The presence of pycnidia and pigmented aseptate, ellipsoidal conidia arising from percurrently proliferating conidiogenous cells were consistent with species placed in Coniothyrium Corda. DNA sequence comparisions have, however, made it possible to recognise that the fungus has a clear phylogenetic position in Mycosphaerella Johanson (Alemu et al. 2005). It is moreover not related to species of Coniothyrium s. str., which are anamorphs of Leptosphaeria spp. This realisation has led to the transfer of Coniothyrium zuluense to Colletogloeopsis Crous & M.J. Wingf. (Cortinas et al. 2006). Colletogloeopsis is a well-recognised Mycosphaerella anamorph and its circumscription was amended to include species with pycnidioid conidiomata. Within Mycosphaerella, C. zuluensis clusters with a group of well-known leaf and stem pathogens of Eucalyptus including M. ambiphylla A. Maxwell, M. cryptica (Cooke) Hansf., M. molleriana (Thüm.) Lindau, M. nubilosa (Cooke) Hansf., M. vespa Carnegie & Keane, M. suttonii Crous & M.J. Wingf., and Phaeophleospora eucalypti (Cooke & Massee) Crous, F.A. Ferreira & B. Sutton (Cortinas et al. 2006).
Different isolates of C. zuluensis have been found to be highly variable in morphology (Fig. 3) and pathogenicity to different Eucalyptus clones (Van Zyl 1997, Wingfield et al. 1997, Van Zyl 2002a). Nonetheless, previous phylogenetic analyses based on the nuclear ribosomal small subunit (18S) and internal transcribed spacer regions and the ribosomal 5.8 gene (ITS1, 5.8S, ITS2) had shown that C. zuluensis was monophyletic (Van Zyl 2002b, Alemu et al. 2005). As additional surveys of Eucalyptus plantations are undertaken, an understanding of the geographical range of C. zuluensis continues to expand. Additional isolates from new regions have thus become available for DNA sequence comparisons and these have provided the opportunity to re-consider the taxonomic status of C. zuluensis, and the variation observed in its morphology and pathogenicity.
Fig. 3.
Characteristics of isolates of Group 1 (C. zuluensis), and isolates of Group 2 (C. gauchensis). Columns A–C show three different colony morphologies belonging to the Group 2 isolates: CMW 7272, CMW 7269, CMW 7293. Columns D–F show three different colony morphologies that belong to the Group 1 isolates: CMW 7488, CMW 5236, CMW 7479.
The aim of this study was to consider whether the previously recognised C. zuluensis can be retained when applying multi-gene analyses using a large collection of isolates not previously available. To accomplish this objective, individual and combined phylogenetic analyses using the ITS region, β-tubulin gene (BT2), the elongation factor 1-α (EF1α) gene, and the mitochondrial ATPase 6 (ATP6) gene, were carried out. Morphological and other phenotypic characters were also considered.
MATERIALS AND METHODS
Isolates
A collection of 45 isolates was chosen to reflect the geographical distribution of C. zuluensis. In addition, several species of Mycosphaerella known to be closely related to C. zuluensis were also included (Table 1). All these isolates were obtained from the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI), Pretoria, South Africa. Single-conidial cultures were established from mature pycnidia isolated from lesions taken from the stems of Eucalyptus trees in South Africa and Uruguay. The contents of single pycnidia were diluted in sterile distilled water, and spread on the surface of Petri dishes containing MEA (20 g/L Biolab malt extract, 15 g/L Biolab agar). After 24–36 h, germinating conidia were transferred to fresh MEA plates and incubated for 30 d at 25 °C. Reference strains are preserved in CMW, and have been deposited at the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands (Table 1). Nomenclature, descriptions and illustrations were deposited in MycoBank.
Table 1.
Isolates of Colletogloeopsis and related species used in the phylogenetic studies.
| Species | Strain numbers | Country | Host | Date |
GenBank no.
|
||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | BT2 | EF1-α | ATP6 | ||||||
| Colletogloeopsis gauchensis | CMW7302 | Uruguay | E. grandis | 2001 | DQ240186 | DQ240075 | DQ240128 | DQ240025 | |
| CMW7274; | CBS 117830 | Uruguay | E.grandis | 2001 | DQ240187 | DQ240076 | DQ240129 | DQ240026 | |
| CMW7294; | CBS 117832 | Uruguay | E. grandis | 2001 | DQ240188 | DQ240077 | DQ240130 | DQ240027 | |
| CMW7300; | CBS 117831 | Uruguay | E. grandis | 2001 | DQ240189 | DQ240078 | DQ240131 | DQ240028 | |
| CMW7270 | Uruguay | E. grandis | 2001 | - | - | - | DQ240068 | ||
| CMW17328 | Uruguay | E. grandis | 2005 | DQ240190 | DQ240079 | DQ240132 | DQ240029 | ||
| CMW17330 | Uruguay | E. grandis | 2005 | DQ240191 | DQ240080 | DQ240133 | DQ240030 | ||
| CMW17323 | Uruguay | E. grandis | 2005 | DQ240215 | DQ240122 | - | DQ240069 | ||
| CMW17324 | Uruguay | E. grandis | 2005 | DQ240216 | DQ240123 | - | DQ240070 | ||
| CMW17326 | Uruguay | E. grandis | 2005 | DQ240217 | DQ240124 | - | DQ240071 | ||
| CMW17332 | Uruguay | E. grandis | 2005 | DQ240218 | DQ240125 | - | DQ240072 | ||
| CMW10895; | CBS 117260 | Hawaii-US | E. grandis | 2002 | DQ240192 | DQ240081 | DQ240134 | DQ240031 | |
| CMW10893; | CBS 117834 | Hawaii-US | E. grandis | 2002 | DQ240193 | DQ240082 | DQ240135 | DQ240032 | |
| CMW10894 | Hawaii-US | E. grandis | 2002 | DQ240194 | DQ240083 | DQ240136 | DQ240033 | ||
| CMW7331; | CBS 117256 | Argentina | E. grandis | 2001 | DQ240195 | DQ240084 | DQ240137 | DQ240034 | |
| CMW7342 | Argentina | E. grandis | 2001 | DQ240196 | DQ240085 | DQ240138 | DQ240035 | ||
| CMW7378 | Argentina | E. grandis | 2001 | DQ240197 | DQ240086 | DQ240139 | DQ240036 | ||
| CMW14336; | CBS 117257 | Argentina | E. grandis | 2003 | DQ240198 | DQ240087 | DQ240140 | DQ240037 | |
| CMW7137 | Uganda | E. grandis | 2001 | DQ240199 | DQ240088 | DQ240141 | DQ240038 | ||
| CMW15835; | CBS 117261 | Uganda | E. grandis | 1999 | DQ240200 | DQ240089 | DQ240142 | DQ240039 | |
| CMW8991; | CBS 117833 | Ethiopia | E. camaldulensis | 2001 | DQ240201 | DQ240090 | DQ240143 | DQ240040 | |
| CMW8978 | Ethiopia | E. camaldulensis | 2001 | DQ240202 | DQ240091 | DQ240144 | DQ240041 | ||
| CMW19356 | Ethiopia | E. camaldulensis | 2000 | - | - | DQ240181 | - | ||
| Colletogloeopsis zuluensis | CMW1772 | South Africa | E. grandis | 1989 | DQ240203 | DQ240092 | DQ240145 | DQ240042 | |
| CMW7426 | South Africa | E. grandis | 1997 | DQ239979 | - | DQ240182 | - | ||
| CMW7459 | South Africa | E. grandis | 1997 | DQ239981 | - | DQ240183 | - | ||
| CMW7488; | CBS 117829 | South Africa | E. grandis | 1997 | DQ239975 | - | DQ240184 | - | |
| CMW7489 | South Africa | E. grandis | 1997 | DQ239980 | - | DQ240185 | - | ||
| CMW17314 | South Africa | E. grandis | 2005 | DQ240204 | DQ240093 | DQ240146 | DQ240043 | ||
| CMW17316 | South Africa | E. grandis | 2005 | DQ240205 | DQ240094 | DQ240147 | DQ240044 | ||
| CMW17320 | South Africa | E. grandis | 2005 | DQ240206 | DQ240095 | DQ240148 | DQ240045 | ||
| CMW17321 | South Africa | E. grandis | 2005 | DQ240207 | DQ240096 | DQ240149 | DQ240046 | ||
| CMW13328; | CBS 113399 | South Africa | E. grandis | - | DQ239974 | - | DQ240172 | - | |
| CMW13324; | CBS 111125 | South Africa | E. grandis | - | AY738214 | - | DQ240173 | - | |
| CMW17318 | South Africa | E. grandis | 2005 | DQ240213 | DQ240126 | DQ240174 | DQ240073 | ||
| CMW17322 | South Africa | E. grandis | 2005 | DQ240214 | DQ240127 | DQ240175 | DQ240074 | ||
| CMW7449; | CBS 117262 | South Africa | E. grandis | 1997 | DQ239976 | DQ240102 | DQ240155 | DQ240052 | |
| CMW7452 | South Africa | E. grandis | 1997 | DQ239977 | DQ240103 | DQ240156 | DQ240053 | ||
| CMW7442 | South Africa | E. grandis | 1997 | DQ239978 | DQ240104 | DQ240157 | DQ240054 | ||
| CMW7468 | South Africa | E. grandis | 1997 | DQ239983 | DQ240105 | DQ240158 | DQ240055 | ||
| CMW15971 | China | E. urophylla | 2004 | DQ240208 | DQ240097 | DQ240150 | DQ240047 | ||
| CMW15080 | China | E. urophylla | 2004 | DQ240209 | DQ240098 | DQ240151 | DQ240048 | ||
| CMW15964 | China | E. urophylla | 2004 | DQ240210 | DQ240099 | DQ240152 | DQ240049 | ||
| CMW17425 | Malawi | E. grandis | 2004 | DQ240211 | DQ240100 | DQ240153 | DQ240050 | ||
| CMW17438 | Malawi | E. grandis | 2004 | DQ240212 | DQ240101 | DQ240154 | DQ240051 | ||
| CMW17356 | Malawi | E. grandis | 2004 | DQ240219 | - | - | - | ||
| CMW6859 | Vietnam | E. urophylla | 2000 | - | DQ240106 | DQ240159 | DQ240056 | ||
| CMW6860 | Vietnam | E. urophylla | 2000 | DQ239985 | DQ240107 | DQ240160 | DQ240057 | ||
| CMW6857; | CBS 118125 | Vietnam | E. urophylla | 2000 | DQ239986 | - | DQ240171 | - | |
| CMW15834; | CBS 117835 | Mexico | E. grandis | 2000 | DQ239987 | DQ240108 | DQ240161 | DQ240058 | |
| CMW15833; | CBS 118149 | Mexico | E. grandis | 2000 | DQ239988 | DQ240109 | DQ240162 | DQ240059 | |
| CMW5235; | CBS 117263 | Thailand | E. camaldulensis | 1997 | DQ239990 | DQ240110 | DQ240163 | DQ240060 | |
| CMW5236 | Thailand | E. camaldulensis | 1997 | DQ239989 | DQ240111 | DQ240164 | DQ240061 | ||
| Mycosphaerella ambiphylla | CMW13704; | CBS 110499 | Australia | Eucalyptus | - | DQ239970 | DQ240116 | DQ240169 | DQ240066 |
| Mycosphaerella colombiensis | CMW4944; | CPC1106 | Colombia | Eucalyptus sp. | - | DQ239993 | DQ240112 | DQ240165 | DQ240062 |
| Mycosphaerella molleriana | CMW4940; | CPC1214 | Portugal | Eucalyptus sp. | 1995 | DQ239969 | DQ240115 | DQ240168 | DQ240065 |
| Mycosphaerella nubilosa | CMW6210; | CBS 114706 | Australia | Eucalyptus sp. | 2000 | DQ239999 | DQ240113 | DQ240166 | DQ240063 |
| Mycosphaerella suttonii | CMW5348; | CPC1346 | Indonesia | Eucalyptus sp. | 1996 | DQ239972 | DQ240117 | DQ240170 | DQ240067 |
| Mycosphaerella vespa | CMW11588 | Australia | Eucalyptus sp. | - | DQ239968 | DQ240114 | DQ240167 | DQ240064 | |
CMW= Culture collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa.
CBS= Culture collection of the Centraalbureau voor Schimmelcultures, Uppsalalaan, Utrecht, The Netherlands. CPC= Culture collection of Pedro Crous housed at CBS.
DNA extraction and amplification
To extract DNA, mycelium was scraped from the surface of cultures grown in Petri dishes, freeze dried, frozen in liquid nitrogen and ground to a fine powder. The protocol followed by Cortinas et al. (2004) was simplified as follows: DBE extraction buffer (200 mM Tris-HCl pH 8, 150 mM NaCl, 25 mM EDTA pH 8, 0.5 % SDS) was added directly to the ground mycelium and incubated for 2 h at 80 °C (or until pigments changed colour from green to red). In the extraction-DNA enrichment procedure, one volume of phenol was used first and one volume of a 1:1 phenol-chloroform solution thereafter.
Four gene regions were amplified for all isolates included in this study (Fig. 4). The ITS region of the ribosomal DNA was targeted using the primers ITS1: 5' TCC GTA GGT GAA CCT GCG G and ITS4: 5' GCT GCG TTC TTC ATC GAT GC (White et al. 1990). Exons 3 to 6 and the respective introns (BT2) of the β-tubulin gene region were amplified using the primers BT2A: 5' GGT AAC CAA ATC GGT GCT GCT TTC and BT2B: 5' AAC CTC AGT GTA GTG ACC CTT GGC (Glass & Donaldson 1995). The intron sequence of the EF1-α gene was amplified using the primers EF1-728F: 5' CAT CGA GAA GTT CGA GAA GG and EF1-986R: 5' TAC TTG AAG GAA CCC TTA CC (Carbone & Kohn 1999) and intron 2 and exon 3 of the ATP6 gene was amplified using the set of primers 5'ATT AAT TSW CCW TTA GAW CAA TT and 5'TAA TTC TAN WGC ATC TTT AAT RTA developed by Kretzer & Bruns (1999).
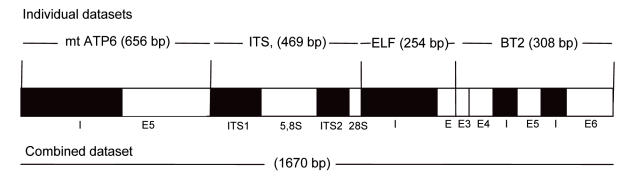
Fig. 4.
Schematic structural organization of the genomic regions used in this study. ITS regions and intron sequences are represented in solid black. Letters “I” indicate introns and letters “E” indicate exons. Sizes of the individual and combined partition alignments are given in brackets. Note that intron between E3 and E4 in the BT2 region is not present.
PCR reactions were prepared in a total volume of 25 μL including 1.5 μL of genomic 1/10 dilution DNA, 1 U of Taq polymerase, 10 × Taq buffer, 10 pmol of each primer, 0.8 mM of each dNTPs, and 2.0 mM MgCl2 (ITS) or 4.0 mM MgCl2 (BT2, EF1-α, ATP6). PCR amplicons were visualised under UV light on 1 % or 2 % agarose gels. Different cycling conditions were used for the various gene regions. For the ITS region, 96 °C, 3 min initial denaturation and cycles of 95 °C, 30 s, 54 °C, 30 s, 72 °C, 1 min were repeated 10 times followed by 25 cycles of 95 °C, 30 s, 56 °C, 30 s, 72 °C, 1 min with 5 s extension after two cycles. A final elongation step of 7 min at 72 °C was also included. The same cycling conditions were used for ATP6 region changing the annealing temperature to 50 °C. For β-tubulin, 96 °C, 3 min initial denaturation and cycles of 95 °C, 30 s, 57 °C, 45 s, 72 °C, 45 s were repeated 40 times. For EF1-α, 96 °C, 3 min and cycles of 95 °C, 30 s, 54 °C, 45 s, 72 °C, 45 s were repeated 40 times with 5 s extension after two cycles. A final elongation step of 7 min at 72 °C was included.
PCR amplification products were purified using Sephadex G-50 columns (Sigma-Aldrich, Steinheim, Germany) or treated with a mix of Exonuclease III and Shrimp alkaline phosphatase (Exo-Sap); 0.7 U of each enzyme per PCR reaction were incubated at 37 °C for 15 min followed by 80 °C for 15 min before sequencing. Sequencing reactions were prepared in 10 μL with 2 μL of purified PCR product, 10 pmol of the same primers used for the first PCR amplifications, 2 μL 5× dilution buffer and ABI Prism Big Dye Terminator mix, v. 3.1 (Applied Biosystems Inc., Foster City, California). Sequencing PCR cycles consisted of 25 repetitions at 96 °C, 10 s; 50 °C, 4 s; 60 °C, 4 min. Sequencing reactions were cleaned using Sephadex G-50 or precipitated using EDTA, Sodium Acetate and Ethanol according to the protocol supplied by Applied Biosystems (Applied Biosystems Inc., Foster City, California).
Phylogenetic analyses
Alignments of sequence data were made using Clustal W under MEGA 3.0 (Kumar et al. 2004) and manually adjusted. All sequences generated in this study were deposited in GenBank (Table 1). Alignments were deposited in TreeBASE.
Maximum parsimony and distance analyses were conducted considering the individual and combined partitions. Most parsimonious (MP) trees were generated using PAUP v. 4.0b10 (Swofford 2002). For parsimony analyses, heuristic searches were used with the steepest descent option and the TBR swapping algorithm. The characters were equally weighted and treated as unordered. Statistical support of the nodes in the trees was tested with 1000 bootstrap replicates. Distance analyses were conducted using MEGA 3.0 (Kumar et al. 2004). Pairwise distances were estimated using the Kimura with two parameters model (Kimura 1980). A gamma distribution α = 0.5 was used to take into account the differences in mutation rate among sites, due to the mix of coding and non-coding sequences present in the analysed fragments. The individual gene reconstructions were performed with Minimum Evolution (Rzhetsky & Nei 1993). Gaps generated in the alignment were treated as missing data. One thousand bootstrap replicates were made to assess the statistical support of the nodes in the phylogenetic trees. Trees were rooted to midpoint.
Partitions were considered together using Bayesian analyses (Ronquist & Huelsenbeck 2003). It has recently been shown that the Bayesian method is more sensitive to under-specification than over-specification of the evolutionary model (Huelsenbeck & Rannala 2004) when calculating the posterior probabilities. Consequently, a time-reversible complex model with gamma-distributed rate variation (GTR + I + G) was selected to combine the data sets. This model of DNA substitution allows the consideration of different rates of substitutions among sites, different nucleotide frequencies, and differences in the rate of substitutions among nucleotides. Therefore, four sets of analyses were run in MrBayes 3.1.1 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003) calculating marginal posterior probabilities using the selected time reversal GTR + I + G model of nucleotide substitution (Tavaré 1986, Yang 1993, 1994) and default values for the prior settings. Four Monte Carlo Markov chains were run for 3 million generations. Trees and parameters were recorded every 100 generations. Likelihood stability was reached at 30 000 generations. This number of generations was then established as the “burn-in” period (represented by 3001 trees). A half compatible consensus tree was recovered from the remaining sampled trees. The Bayesian procedure was repeated four times. The posterior probabilities are indicated close to the respective nodes on the tree and the sequences of Mycosphaerella colombiensis Crous & M.J. Wingf. and M. suttonii Crous & M.J. Wingf. were used as outgroups.
Temperature sensitivity studies
Plugs (3 mm diam) of colonised agar were cut from actively growing cultures and placed at the centres of Petri dishes containing MEA. Isolates tested for growth characteristics at different temperatures included those from South Africa (CMW 7442, CMW 7449, CMW 7479, CMW 7488), and others from Uruguay (CMW 7269, CMW 7274, CMW 7279, CMW 7300). Three plates were prepared for each isolate and these were incubated at temperatures between 5 °C and 35 °C at 5 ° intervals, for 6 wk. A second set of isolates from Ethiopia (CMW 8282, CMW 8292) and from China (CMW 15966, CMW 15971) were tested in a similar manner but for an incubation period of 8 wk. Growth was recorded weekly by measuring average colony diameter.
Morphology
Descriptions are based on sporulation in vivo. Wherever possible, 30 measurements (× 1000 magnification) were made of structures mounted in lactic acid, the 95 % deviation determined, and the extremes of spore measurements given in parentheses. Colony colours (surface and reverse) were assessed after 25 d on MEA at 25 °C in the dark, using the colour charts of Rayner (1970).
RESULTS
PCR and sequence analyses
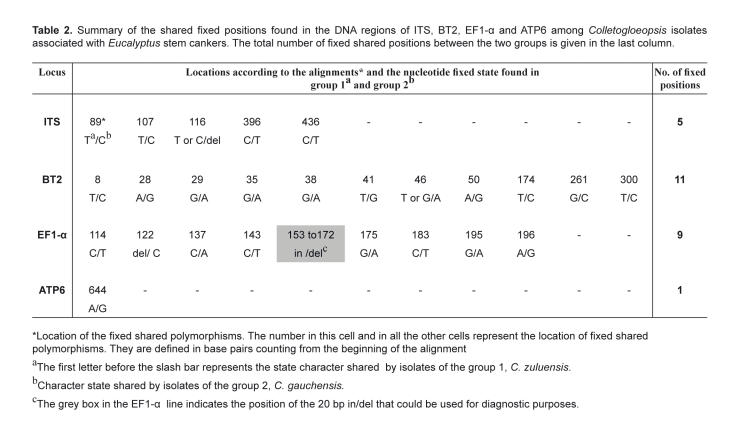
Sequenced amplicons obtained from C. zuluensis isolates for the four different gene regions were aligned to study fixed polymorphisms. Alignments of 469 bp (ITS), 308 bp (BT2), 254 bp (EF1-α) and 656 bp (ATP6) were generated. The intron between the exons 3 and 4 of the β-tubulin gene was missing in all isolates studied. Visual analyses of the characters defined two groups among the isolates based on the fixed, shared polymophisms. The first group included isolates from South Africa, China, Thailand, Vietnam and Malawi and a second group comprised isolates from Uruguay, Argentina, Hawaii, Uganda and Ethiopia. Positions in base pairs of the different fixed characters in the alignments for the various isolates are shown in Table 2. Five fixed characters were found at the ITS region, eleven were found in the BT2 dataset, eight were found at the EF1-α intron where a 20-base-pair indel was also found (Fig. 5). One fixed position was found in the ATP6 region.
Table 2.
Summary of the shared fixed positions found in the DNA regions of ITS, BT2, EF1-α and ATP6 among Colletogloeopsis isolates associated with Eucalyptus stem cankers. The total number of fixed shared positions between the two groups is given in the last column.
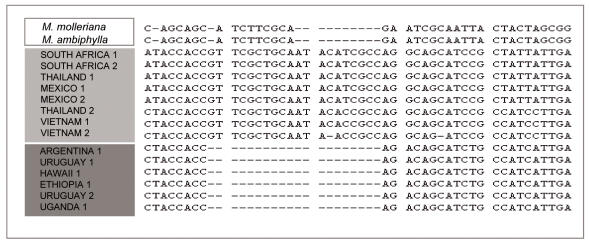
Fig. 5.
Partial alignment of isolates showing the characteristic 20 bp elongation factor 1-α in/del. The presence of the in/del identifies the Group 1 isolates (light grey) from Group 2 (dark grey) isolates. All isolates in Table 1 can be assigned correctly into Groups 1 or 2 according to the presence/absence of this fragment.
Phylogenetic analyses
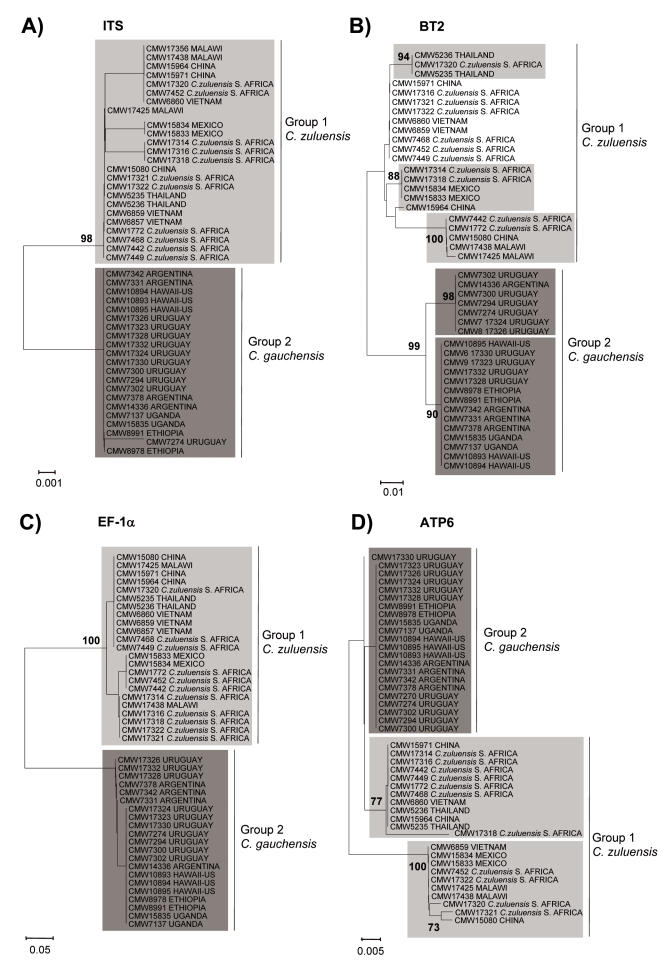
Individual phylograms were obtained for each gene region and parsimony data produced very similar topologies to those of the distance trees. Therefore, only distance trees are presented (Fig. 6). In all cases the Bootstrap cut-off of 70 % was established.
Fig. 6.
Phylograms generated using Minimum Evolution and K2P with gamma distribution, α= 1. A. ITS. B. β-tubulin. C. EF1-α. D. ATP6. Values on branches are bootstrap support (1000 replicas).
Analyses of sequence data for the ITS region resolved two coherent clusters for the Colletogloeopsis isolates considered. These groups represented isolates from South Africa, Malawi, Mexico, Thailand, Vietnam and China (Group1) and those from Uruguay, Argentina, Hawaii, Ethiopia and Uganda (Group 2). The separation of these two groups had 98 % bootstrap support in the ITS tree. In the BT2 and EF-1α trees, these two groups had 99 % and 100 % support, respectively. For the ATP6 tree, three groups could be distinguished although only one of these had strong support (100 %). The group having reasonable support included isolates from Vietnam, Mexico, Malawi, China and South Africa. Internal sub-clusters could be distinguished within the Group 1 and Group 2 clusters in the ITS, BT2 and EF1-α trees. These sub-clusters had greater than 70 % bootstrap support only in the BT2 tree. The assortment of isolates within the sub-clusters was different in different trees.
The level of polymorphism observed in the datasets was different for each individual analysed region. The β-tubulin data set presented the highest level of variation followed by the EF1-α, ITS and ATP6 data sets, respectively. A close inspection of the ATP6 data matrix showed few polymorphisms explaining the poor resolution obtained in the tree.
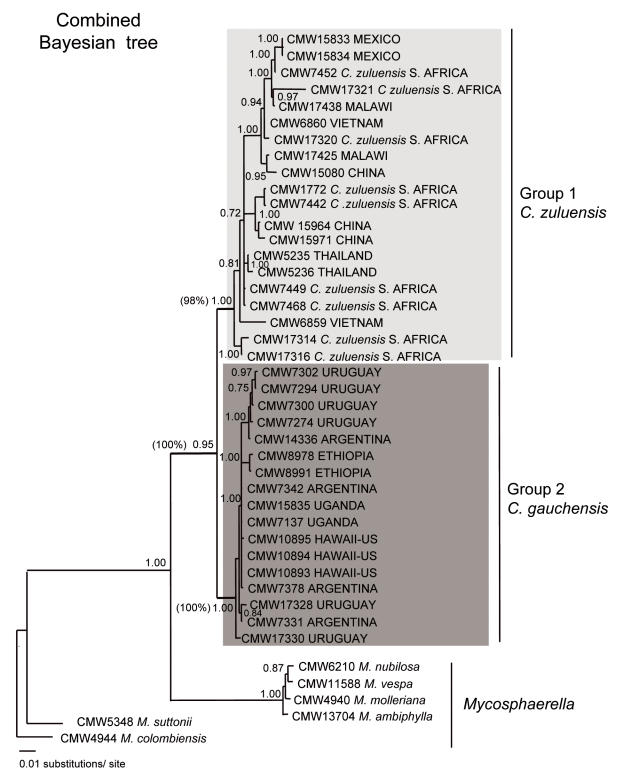
After the individual analyses, combined parsimony and Bayesian analysis were carried out (Fig. 7). The reconstructed trees included the collection of Colletogloeopsis isolates together with Mycosphaerella spp. A posterior probability of 1 and a 100 % bootstrap value separated the Colletogloeopsis isolates from the rest of Mycosphaerella spp. The parsimony and Bayesian half-compatible trees showed two major groups representing isolates from South Africa, Malawi, Mexico, Thailand, Vietnam and China (Group1) and those from Uruguay, Argentina, Hawaii, Ethiopia and Uganda (Group 2) supported by posterior probabilities of 1 and 0.95 and 98 % and 100 % bootstrap values, respectively. A rich internal topology was found within these two groups. Numerous sub-clusters were supported with high probabilities and bootstrap values. A number of these included more than one isolate from the same locality. Nevertheless, location was not sufficient to explain how the sub-clusters were formed.
Fig. 7.
Bayesian combined tree using a GTR+G+I model of substitutions. Posterior probabilities are shown on the branches. Parsimony bootstrap values are shown in brackets.
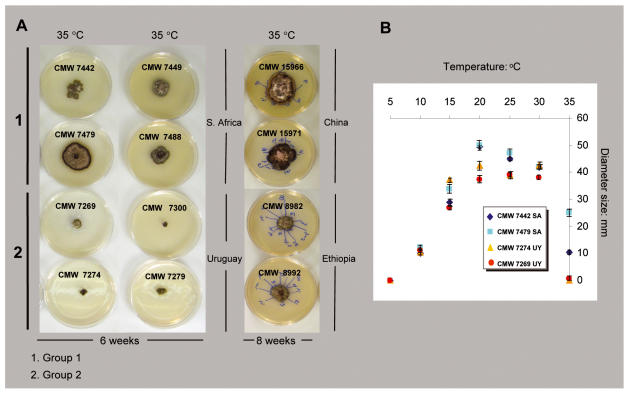
Temperature sensitivity studies
Average colony diameter for the isolates from South Africa and from Uruguay was different at some of the tested temperatures after 6 wk (Fig. 8). No measurable growth was found at 5 °C, optimal growth occurred between 20 and 25 °C, and the diameters of colonies decreased when they were incubated at temperatures of 30 °C and above. Differences between isolates from the two regions were seen at 10 °C where the Uruguayan isolates grew more rapidly than isolates from South Africa. Between 20 °C and 25 °C both groups of isolates achieved their maximum diameter. Nevertheless, these maximum diameters were smaller for the Uruguayan isolates. The most obvious difference between South African and Uruguayan isolates was observed at 35 °C. At this temperature, the Uruguayan isolates hardly displayed growth whereas South African isolates reached between 10 and 20 mm diam.
Fig. 8.
Results of culture growth studies at different temperatures. A. Isolates from South Africa and Uruguay were tested for a period of 6 weeks and those from China and Ethiopia for a period of 8 weeks. Each point on the graph represents the average of 6 measurements taken at each temperature.
The results obtained in a second experiment including isolates from China and Ethiopia, were very similar to those comparing isolates from South Africa and Uruguay. After 8 wk, the differences in growth of the isolates from both origins were obvious at 35 °C (Fig. 8). This is consistent with the fact that isolates from China are phylogenetically related to those from South Africa and those from Ethiopia are related to those from Uruguay.
Morphology
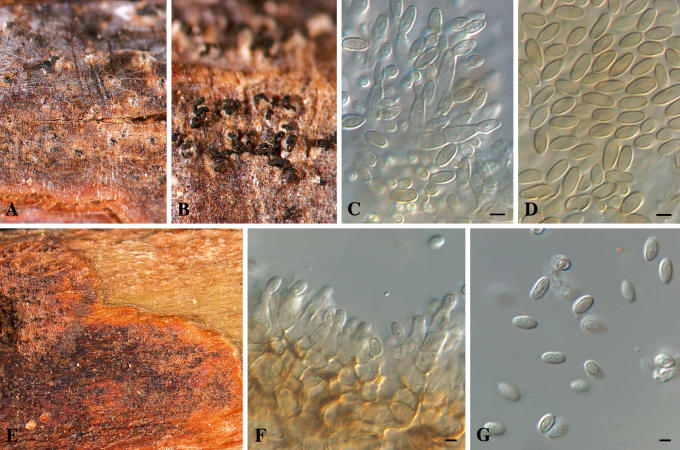
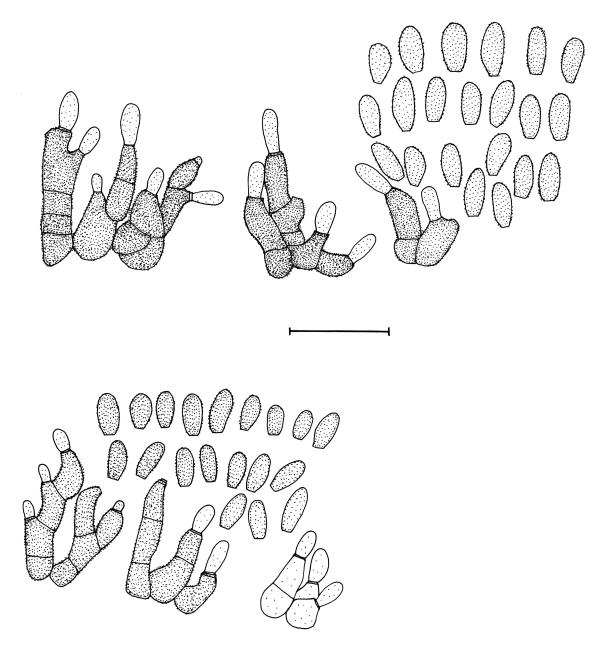
Isolates of Colletogloeposis included in this study were morphologically variable in culture. Colony characteristics overlapped for isolates from South Africa and Uruguay, but it was possible to recognise some characteristics apparently exclusive to the Uruguayan isolates. Likewise, distinctly different conidial and conidiogenous cell characteristics were found when isolates from Uruguay were compared with those of C. zuluensis from South Africa (Fig. 9). The range of conidial lengths overlapped almost entirely between C. zuluensis [conidia (4–)4.5–5(–6) × 2–2.5(–3.5) μm] and the isolates from Uruguay [conidia (4–)5–6(–7.5) × (2–)2.5(–3) μm]. The Uruguayan conidia, however, had a larger maximum length, reaching 7.5 μm (6 μm for C. zuluensis). Conidia of C. zuluensis were slightly wider (3.5 μm) as opposed to those from Uruguay, which were an average of 3 μm. Another distinctive characteristic of the fungus from Uruguay is that it has sympodial polyphialidic conidiogenous cells, which is different to C. zuluensis, which has percurrently proliferating monophialidic conidiogenous cells.
Fig. 9.
Colletogloeopsis spp. sporulating on E. grandis stems. A–D. Colletogloeopsis gauchensis (holotype). A–B. Pycnidia with black cirri. C. Conidiogenous cells. D. Conidia. E–G. Colletogloeopsis zuluensis (epitype). E. Pycnidia. F. Conidiogenous cells. G. Conidia. Scale bars = 2.5 μm.
Taxonomy
Phylogenetic analyses in this study supported two distinct groups of isolates, encompassed within the fungus currently treated as C. zuluensis. One of these groups of isolates is from South Africa, Malawi, Thailand, Vietnam, China and Mexico. The other group includes isolates from Uruguay, Argentina, Hawaii-U.S.A., Ethiopia and Uganda. These fungi can also be separated by characteristics of growth in culture, morphology and growth at different temperatures. Clearly, the South African fungus must retain the name C. zuluensis. At the time of describing this fungus, no ex-type cultures were deposited. We have thus provided a suite of isolates for which DNA sequence data are available, and that are tied to herbarium specimens to serve as epitypes. The fungus occurring in Uruguay and other countries represents a distinct taxon that is described below.
Colletogloeopsis gauchensis M.-N. Cortinas, Crous & M.J. Wingf., sp. nov. MycoBank MB500854. Figs 9, 10.
Fig. 10.
Colletogloeopsis spp. sporulating on E. grandis stems. Conidiogenous cells and conidia of Colletogloeopsis gauchensis (holotype) (top). Conidiogenous cells and conidia of Colletogloeopsis zuluensis (epitype) (bottom). Scale bar = 10 μm.
Etymology: Named after the gauchos people of South America that live in the same area where this species is distributed and where it was first collected. In the same genus, C. zuluensis is named after the KwaZulu-Natal Province and the “Zulu” people of South Africa.
Colletogloeopsidi zuluensi similis, sed conidiis angustioribus, (4–)5–6(–7.5) × (2–)2.5(–3) μm et phialidibus nonnumquam sympodialiter proliferentibus distincta.
Lesions caulicolous, subcircular to irregular, dark brown, 2–10 mm diam, with a raised, red-brown border. Conidiomata pycnidial to somewhat acervular, subepidermal, single, rarely aggregated, occurring in necrotic tissue, globose to slightly depressed, becoming erumpent, up to 120 μm diam, exuding conidia in a long cirrus; conidiomatal walls composed of 2–3 layers of medium brown textura angularis; opening by a central ostiole or irregular rupture; ostiolar region lined with thick-walled, brown, smooth, septate hyphae that are sometimes branched below, 3–4 μm wide, with obtuse ends that flare apart (upper 1–6 cells). Conidiophores subcylindrical, subhyaline to medium brown, smooth to finely verruculose, 0–3-septate, unbranched or branched below, 10–20 × 3–6 μm. Conidiogenous cells subhyaline to medium brown, dolilform to subcylindrical, smooth to finely verruculose, mono- to polyphialidic, proliferating percurrently, with several percurrent proliferations near the apex. Conidia medium brown, thick-walled, finely verruculose, broadly ellipsoidal, apex obtuse to subobtuse, base subtruncate to bluntly rounded, (4–)5–6(–7.5) × (2–)2.5(–3) μm; base frequently with a minute marginal frill.
Specimens examined: Uruguay, El Tarugo, bark of 1-yr-old E. grandis tree, Feb. 2005, M.J. Wingfield, CBS H-19724 holotype, cultures ex-holotype CMW 17331–17332; La Herradura, CBS H-19722, cultures CBS 119467-119466 = CMW 17542–17543; ibid., CBS H-19723, cultures CBS 119465 = CMW 17545, CMW 17544; La Juanita, CBS H-19725, cultures CBS 119468 = CMW 17558, CMW 17559; ibid., CBS H-19726, cultures = CMW 17560–17561.
Cultural characteristics: Colony characteristics on MEA at 25 °C are variable. Colony colours were similar to those of C. zuluensis (Van Zyl et al. 1997, 2002). Surface colours range from greyish yellow-green, dull green, isabelline, greenish olivaceous to grey-olivaceous; colonies in reverse range from dark grey, dark olive-grey to dark green (Rayner 1970); margins are smooth, regular or irregular. Some cultures develop a characteristic white outer zone of aerial mycelium (Fig. 3). Paler colonies develop smoother surfaces with white aerial mycelium; some strains produce a diffuse yellow pigment in MEA.
Notes: Colletogloeopsis gauchensis [conidia (4–)5–6(–7.5) × (2–)2.5(–3) μm] can readily be distinguished from C. zuluensis [conidia (4–)4.5–5(–6) × 2–2.5(–3.5) μm] by its slightly longer conidia, and the presence of sympodial polyphialidic conidiogenous cells (Figs 9, 10). Furthermore, it grows readily at 10 °C, with hardly any to no growth at 35 °C. In contrast, C. zuluensis grows more slowly at 10 °C, and faster at 35 °C than C. gauchensis, and strains of C. gauchensis do not form conidiomata in culture.
Colletogloeopsis zuluensis (M.J. Wingf., Crous & T.A. Cout.) M.-N. Cortinas, M.J. Wingf. & Crous, Mycol. Res. 110: 235. 2006. Figs 9, 10. [as zuluense].
Basionym: Coniothyrium zuluense M.J. Wingf., Crous & T.A. Cout., Mycopathologia 136: 142. 1997.
Specimens examined: South Africa, KwaZulu-Natal, Kwambonambi, Teza nursery, bark of 1-yr-old E. grandis tree, Jan. 1996, M.J. Wingfield, IMI 370886 holotype; KwaZulu-Natal, Kwambonambi, E. grandis, Feb. 2005, M.J. Wingfield, CBS H-19721 epitype here designated, culture ex-epitype CMW 17321–17322; CBS H-19717, culture CBS 119427 = CMW 17531, CMW 17530; CBS H-19720, culture CBS 119471 = CMW 17528, CMW 17529; CBS H-19719, culture CBS 119470 = CMW 17320, CMW 17319; CBS H-19718, culture CBS 119469 = CMW 17526, CMW 17527.
DISCUSSION
Phylogenetic analyses for a large number of C. zuluensis isolates from different parts of the world and based on multiple gene regions have shown clearly that this material represents at least two discrete taxa. These species are described based on material from South Africa and Uruguay, but both taxa include collections from many different countries. Thus C. zuluensis is now known from South Africa, Malawi, Thailand, Vietnam, China and Mexico. Likewise, C. gauchensis described in this study occurs not only in Uruguay but also in Argentina, Hawaii-U.S.A., Ethiopia and Uganda. The two fungi thus represent distinct phylogenetic species but they can clearly be distinguished from each other based on morphological characteristics and growth characteristics in culture.
Twenty-six fixed nucleotide positions allowed us to separate the collection of C. zuluensis s. lat. isolates used in this study into two distinctive groups. One of these fixed polymorphisms found in the EF1-α intron can easily be used to discriminate between C. zuluensis and C. gauchensis. This 20 bp fragment between positions 153 to 172 in C. zuluensis is absent in C. gauchensis. The p-distance among the Colletogloeopsis isolates considered in this study displayed a range of 0 to 1 % divergence in ITS sequences, 0–8 % for BT2 sequences, 0–24 % for EF1- α sequences and 0–4 % for ATP6 data-matrices respectively. These ranges showed that there was sufficient variation within Colletogloeopsis to suspect that more than one taxon was represented in the collection of isolates. The distances are also consistent with values used in previous studies (Couch & Kohn 2002, Barnes et al. 2005) to separate taxa.
Very few morphological differences were found between isolates of C. zuluensis from South Africa and isolates of C. gauchensis from Uruguay. These differences include the fact that Uruguayan isolates have polyphialidic, sympodially and percurrently proliferating conidiogenous cells as opposed to the monophialidic, percurrently proliferating conidiogenous cells in C. zuluensis. The conidia of C. gauchensis are also consistently longer than those of C. zuluensis (Figs 9, 10). Furthermore, C. gauchensis is adapted to cooler climates than C. zuluensis. On the contrary, isolates of C. zuluensis grow well at 35 °C, whereas those of C. gauchensis barely grow at this temperature.
Results of this study provide added support for the view that C. zuluensis and C. gauchensis are anamorphs of Mycosphaerella. They have an allopatric distribution and are considered sibling species only in terms of the fact that they are ecologically and morphologically very similar. The extent to which cryptic and sibling species occur in taxonomic groups varies depending on the group of fungi studied. However, the discovery of cryptic species such as C. gauchensis in this study is becoming a commonplace when DNA studies are implemented (see Crous et al. 2006). Results of such studies reveal that these species reflect collections of morphologically similar taxa that can only be discriminated based on minute morphological details or characteristics in pure culture. A further example of such a species complex in Mycosphaerella concerns “Coniothyrium” ovatum H.J. Swart (Crous et al. 2004a, b, 2006 – this volume), which will be treated elsewhere (Cortinas et al. in prep.).
Intraspecific variation detected amongst isolates of C. zuluensis and to a lesser extent C. gauchensis showed internal structure in the individual and combined trees. Such intraspecific structure was only well-supported in the BT2, ATP6 and combined trees. Based solely upon the phylogenetic species concept, it would be possible to recognise additional species especially in this complex. For the present, however, we choose to not provide additional names before robust population biology studies are available.
Coniothyrium canker is one of the most important diseases of Eucalyptus worldwide (Old et al. 2003). In South Africa, it appeared relatively suddenly in a very limited location and spread rapidly, resulting in very substantial losses to the local forestry industry. The disease has also caused substantial damage to plantations in other countries such as Argentina and Uruguay. It is thus intriguing that there are two distinct fungi associated with indistinguishable symptoms. The origin of the fungus is unknown and it is not known to occur in the native range of Eucalyptus. The evidence from this study shows that the two fungi are closely related and have adapted differently based on some ecological factor. Like most Mycosphaerella spp. they are highly host-specific to certain species of Eucalyptus, grow poorly in culture, and thus it seems reasonable to expect that their origin would be on Eucalyptus or a host closely related to it. A similar situation has emerged for species of Chrysoporthe Gryzenh. & M.J. Wing. (Gryzenhout et al. 2004). that are well-known pathogens of Eucalyptus but that appear to have originated on a wide variety of woody plants in the order Myrtales (Wingfield 2003, Gryzenhout et al. 2004, Seixas et al. 2004).
Recognition of two species within a collection of isolates that have previously been recognised as belonging to the single taxon has important consequences for disease control and quarantine. In the past, it has been suggested that the fungus originated in South Africa, and that it was restricted to that country (Wingfield et al. 1997). Thus, the appearance of the disease in other countries has often been linked to the movement of plant material and particularly seed to other countries. Although it has not been shown experimentally that C. zuluensis is moved on seed, this appears to be a likely mode of global distribution. There is a large international trade in Eucalyptus seed, which is variably controlled and monitored. Both C. zuluensis and C. gauchensis have now wide geographic distributions and this implies that they have been spread from one or a number of sources. Every effort should now be made to restrict them from further movement to new countries and areas.
Acknowledgments
We thank the FABI administrative and culture collection support staff as well as our colleagues Irene Barnes, Wolfgang Maier and Gavin Hunter for their assistance and helpful comments. We also acknowledge the National Research Foundation, members of the Tree Protection Co-operative Program (TPCP), the Centre of Excellence in Tree Health Biotechnology and the THRIP initiative of the Department of Trade and Industry, South Africa for financial support.
Taxonomic novelty: Colletogloeopsis gauchensis M.-N. Cortinas, Crous & M.J. Wingf. sp. nov.
References
- Alemu G, Cortinas MN, Wingfield MJ, Roux J (2005). Characterisation of the Coniothyrium stem canker pathogen on Eucalyptus camaldulensis in Ethiopia. Australasian Plant Pathology 34: 1–6. [Google Scholar]
- Alemu G, Roux J, Thu PQ, Wingfield MJ (2004). Coniothyrium stem canker of Eucalyptus, new to Argentina and Vietnam. South African Journal of Science 99: 587–588. [Google Scholar]
- Alemu G, Roux J, Wingfield MJ (2003). Diseases of exotic plantation Eucalyptus and Pinus species in Ethiopia. South African Journal of Science 99: 29–33. [Google Scholar]
- Barnes I, Crous PW, Wingfield BD, Wingfield MJ (2004). Multigene phylogenies reveal that red band needle blight of Pinus is caused by two distinct species of Dothistroma, D. septosporum and D. pini. Studies in Mycology 50: 551–565. [Google Scholar]
- Carbone I, Kohn LM (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Cortinas MN, Burgess T, Dell B, Xu D, Crous PW, Wingfield BD, Wingfield MJ (2006). First record of Colletogloeopsis zuluensis comb. nov., causing stem canker of Eucalyptus in China. Mycological Research 110: 229–236. [DOI] [PubMed] [Google Scholar]
- Cortinas MN, Koch N, Thain J, Wingfield BD, Wingfield MJ (2004). First record of the Eucalyptus stem canker pathogen, Coniothyrium zuluense from Hawaii. Australasian Plant Pathology 33: 309–312. [Google Scholar]
- Couch BC, Kohn LM (2002). A multilocus gene genealogy concordant with host preferences indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 94: 683–693. [DOI] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Mansilla JP, Hunter GC, Wingfield MJ (2004a). Phylogenetic reassessment of Mycosphaerella species and their anamorphs occurring on Eucalyptus. Studies in Mycology 50: 195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Pongpanich K, Himaman W, Arzanlou M, Wingfield MJ (2004b). Cryptic speciation and host specificity among Mycosphaerella species occurring on Australian Acacia species grown as exotics in the tropics. Studies in Mycology 50: 457–469. [Google Scholar]
- Crous PW, Wingfield MJ, Mansilla JP, Alfenas AC, Groenewald JZ (2006). Phylogenetic reassessment of Mycosphaerella species and their anamorphs occurring on Eucalyptus. II. Studies in Mycology 55: 99–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryzenhout M, Myburg H, Merwe NA van der, Wingfield BD, Wingfield MJ (2004). Chrysoporthe, a new genus to accommodate Cryphonectria cubensis. Studies in Mycology 50: 119–142. [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001). MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Rannala B (2004). Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Systematic Biology 53: 904–913. [DOI] [PubMed] [Google Scholar]
- Kimura M (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kretzer A, Bruns TD (1999). Use of ATP6 in fungal phylogenetics: An example from Boletales. Molecular Phylogenetics and Evolution 13: 483–492. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Old KM, Wingfield MJ, Yuan ZQ (2003). A manual of diseases of eucalypts in South-East Asia, Center for International Forestry Research, Bogor, Indonesia.
- Rayner, R. W. (1970). A mycological colour chart. Commonwealth Mycological Institute, Kew.
- Roux J, Wingfield MJ, Cibrián D (2002). First report of Coniothyrium canker in Mexico. Plant Pathology 51: 382. [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003). MrBayes 3.0: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A, Nei M (1993). Theoretical foundation of the minimum-evolution method of phylogenetic inference. Molecular Biology and Evolution 10: 1073–1095. [DOI] [PubMed] [Google Scholar]
- Seixas CDS, Barreto RW, Alfenas AC, Ferreira FA (2004). Cryphonectria cubensis on an indigenous host in Brazil: a possible origin for eucalyptus canker disease? Mycologist 18: 18–24. [Google Scholar]
- Slippers B, Stenlid J, Wingfield MJ (2005). Emerging pathogens: fungal host jumps following anthropogenic evolution. Trends in Ecology and Evolution 20: 420–421. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002). PAUP*: Phylogenetic Analysis using Parsimony (*and other methods). Version 4. Sinauer Associates. Sunderland, Massachusetts, U.S.A.
- Tavaré S (1986). Some probabilistic and statistical problems on the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences 17: 57–86. [Google Scholar]
- Van Zyl LM, Coutinho TA, Wingfield MJ (2002a). Morphological, cultural and pathogenic characteristics of Coniothyrium zuluense isolates from different plantation regions in South Africa. Mycopathologia 155: 149–153. [DOI] [PubMed] [Google Scholar]
- Van Zyl LM, Coutinho TA, Wingfield MJ, Pongpanich K, Wingfield BD (2002b). Morphological and molecular relatedness of geographically diverse isolates of Coniothyrium zuluense from South Africa and Thailand. Mycological Research 106: 51–59. [Google Scholar]
- Van Zyl LM, Wingfield MJ, Coutinho TA (1997). Diversity among isolates of Coniothyrium zuluense, a newly recorded Eucalyptus stem canker pathogen in South Africa. In: Proceedings of the IUFRO Conference on Silviculture and Improvement of Eucalyptus (EMBRAPA/CNPF). Salvador, 24–29 August 1997, Brazil. Anais, Colombo. 3: 135–141. [Google Scholar]
- White TJ, Bruns T, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylognetics. In: A Guide to Molecular Methods and Applications (Innis MA, Gelfand DH, Sninsky JJ, White JW, eds). Academic Press, New York: 315–322.
- Wingfield MJ (2003). Increasing threat of diseases to exotic plantation forests in the southern hemisphere: Lessons from Cryphonectria canker. Australasian Plant Pathology 32: 133–139. [Google Scholar]
- Wingfield MJ, Crous PW, Coutinho TA (1997). A serious new canker disease of Eucalyptus in South Africa caused by a new species of Coniothyrium. Mycopathologia 136: 139–145. [DOI] [PubMed] [Google Scholar]
- Yang Z (1993). Maximum likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Molecular Biology and Evolution 10: 1396–1401. [DOI] [PubMed] [Google Scholar]
- Yang Z (1994). Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. Journal of Molecular Evolution 39: 306–314. [DOI] [PubMed] [Google Scholar]