Abstract
The genus Quambalaria consists of plant-pathogenic fungi causing disease on leaves and shoots of species of Eucalyptus and its close relative, Corymbia. The phylogenetic relationship of Quambalaria spp., previously classified in genera such as Sporothrix and Ramularia, has never been addressed. It has, however, been suggested that they belong to the basidiomycete orders Exobasidiales or Ustilaginales. The aim of this study was thus to consider the ordinal relationships of Q. eucalypti and Q. pitereka using ribosomal LSU sequences. Sequence data from the ITS nrDNA were used to determine the phylogenetic relationship of the two Quambalaria species together with Fugomyces (= Cerinosterus) cyanescens. In addition to sequence data, the ultrastructure of the septal pores of the species in question was compared. From the LSU sequence data it was concluded that Quambalaria spp. and F. cyanescens form a monophyletic clade in the Microstromatales, an order of the Ustilaginomycetes. Sequences from the ITS region confirmed that Q. pitereka and Q. eucalypti are distinct species. The ex-type isolate of F. cyanescens, together with another isolate from Eucalyptus in Australia, constitute a third species of Quambalaria, Q. cyanescens (de Hoog & G.A. de Vries) Z.W. de Beer, Begerow & R. Bauer comb. nov. Transmission electron-microscopic studies of the septal pores confirm that all three Quambalaria spp. have dolipores with swollen lips, which differ from other members of the Microstromatales (i.e. the Microstromataceae and Volvocisporiaceae) that have simple pores with more or less rounded pore lips. Based on their unique ultrastructural features and the monophyly of the three Quambalaria spp. in the Microstromatales, a new family, Quambalariaceae Z.W. de Beer, Begerow & R. Bauer fam. nov., is described.
Keywords: Cerinosterus, Fugomyces, ITS, LSU, Microstromatales, Sporothrix, Ramularia, ultrastructure, Ustilaginomycetes
INTRODUCTION
During the 1950's, a shoot disease was observed on Corymbia maculata (then Eucalyptus maculata) seedlings in New South Wales, Australia. The causal fungus was later described as Ramularia pitereka J. Walker & Bertus (Walker & Bertus 1971). In 1987, a similar disease was noted on a Eucalyptus grandis clone in South Africa. Wingfield et al. (1993) described the South African fungus as a new species, Sporothrix eucalypti M.J. Wingf., Crous & Swart. In his monograph of Ramularia Unger, Braun (1998) transferred R. pitereka to Sporothrix Hektoen & C.F. Perkins. In the same volume, a third Sporothrix species, S. pusilla U. Braun & Crous, isolated from leaf spots on Eucalyptus camaldulensis in Thailand, was described. Braun (1998) distinguished the three species based on morphology and host specificity. The treatment of the three species in Sporothrix (Ophiostomataceae, Ophiostomatales), and not Ramularia (Mycosphaerellaceae, Mycosphaerellales), was based largely on conidial scar morphology (Braun 1998).
Studies prior to Braun's (1998) treatment of the Eucalyptus pathogens as species of Sporothrix had shown that this genus accommodates superficially similar species with diverse phylogenetic relationships (Weijman & De Hoog 1985, De Hoog 1993). The type species for the genus Sporothrix, S. schenckii Hekt. & C.F. Perkins, was placed in the teleomorph genus Ophiostoma Syd. & P. Syd., based on 18S rDNA sequences (Berbee & Taylor 1992). More recently, Simpson (2000) showed that isolates of R. pitereka are not cycloheximide-tolerant, as is almost always the case with Sporothrix isolates with affinities to Ophiostoma (Harrington 1981). Based on the cycloheximide intolerance, pathogenicity to species of Eucalyptus and Corymbia, the dense growth of white conidiophores on agar media and the host, and the absence of distinct denticles on the conidiogenous cells, Simpson (2000) concluded that the affinities of R. pitereka and the two related species, S. eucalypti and S. pusilla, are not with the Ophiostomataceae. He consequently erected the new genus, Quambalaria J.A. Simpson, to accommodate the three species. Simpson (2000), like Braun (1998), distinguished the species based on conidial morphology and specificity to their respective Eucalyptus or Corymbia hosts. Furthermore, based on the apparent absence of dolipore septa in their hyphae observed by light microscopy, he suggested that these fungi probably reside in either one of the basidiomycete orders Exobasidiales Henn., emend. R. Bauer & Oberw., or Ustilaginales G. Winter, emend. R. Bauer & Oberw. (Simpson 2000).
There had been one other Sporothrix-like fungus isolated from Eucalyptus pauciflora in Australia by V.F. Brown. This isolate was sent to CBS in 1973 and was identified as Sporothrix cyanescens de Hoog & G.A. de Vries, earlier described from human skin (De Hoog & De Vries 1973). Smith & Batenburg-Van der Vegte (1985) confirmed that S. cyanescens, and also S. luteoalba de Hoog, have dolipores in their septa and are thus the anamorphs of basidiomycetes. Based on this fact and the presence of the basidiomycetous coenzyme Q-10 system (Suzuki & Nakase 1986), Moore (1987) erected a new genus, Cerinosterus R.T. Moore, for the two Sporothrix spp., with C. luteoalbus (de Hoog) R.T. Moore as generic type species. The first phylogenetic study that included the two Cerinosterus spp. showed that C. luteoalbus groups within the Dacrymycetales Henn. based on LSU sequences (Middelhoven et al. 2000). However, C. cyanescens (de Hoog & G.A. de Vries) R.T. Moore grouped in the Microstromatales R. Bauer & Oberw., and it was suggested that it could not be accommodated in Cerinosterus. Sigler & Verweij (2003) thus described a new genus, Fugomyces Sigler, with F. cyanescens (de Hoog & G.A. de Vries) Sigler as type species.
The aim of this study was to determine whether Quambalaria spp. are monophyletic and what their relationship was to F. cyanescens, using ITS sequences. Furthermore, ribosomal LSU sequences and ultrastructural characters were used to determine an appropriate order in which species of Quambalaria should reside.
MATERIALS & METHODS
Isolates and herbarium specimens
For phylogenetic studies, two South African isolates of Q. eucalypti (M.J. Wingf., Crous & W.J. Swart) J.A. Simpson, including the ex-type culture (CMW 1101 = CBS 118844), were compared with two isolates representing Q. pitereka (J. Walker & Bertus) J.A. Simpson from recent disease outbreaks in Queensland, Australia (Table 1). Two isolates representing F. cyanescens, including the ex-type culture (CBS 357.73), were also included. Other isolates for which DNA sequences were obtained in this study, are listed in Table 1. GenBank accession numbers of sequences obtained in previous studies, are indicated in Figs 1, 2.
Table 1.
Isolates and herbarium specimens used in this study.
| Species | CBS numbers | Isolate number | Herbarium number | Host | Origin | Collector |
GenBank
|
|
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | |||||||
| Microstroma album | R.B. 2072 | Quercus robur | Germany | R. Bauer | DQ317624 | AF352052 | ||
| M. juglandis | F3381 | Jugians regia | Germany | M. Göker | DQ317632 | — | ||
| R.B. 2054 | J. regia | Germany | R. Bauer | DQ317633 | — | |||
| R.B. 2042 | J. regia | Germany | R. Bauer | DQ317634 | DQ317617 | |||
| Quambalaria cyanescens = Fugomyces cyanescens | CBS 357.731T | CMW 5583 | skin of man | Netherlands | T.F. Visser | DQ317622 | DQ317615 | |
| CBS 876.73 | CMW 5584 | Eucalyptus pauciflora | New South Wales, Australia | V.F. Brown | DQ317623 | DQ317616 | ||
| Q. eucalypti | CBS 118844T | CMW 1101 | PREM 51089T | E. grandis | Kwambonambi, South Africa | M.J. Wingfield | DQ317625 | DQ317618 |
| CBS 119680 | CMW 11678 | PREM 58939E | E. grandis clone NH58 | Kwambonambi, South Africa | L. Lombard | DQ317626 | DQ317619 | |
| Q. pitereka | CMW 6707 | Corymbia maculata | New South Wales, Australia | M.J. Wingfield | DQ317627 | DQ317620 | ||
| CBS 118828 | CMW 5318 | C. citriodora subsp. variegata | Queensland, Australia | M. Ivory | DQ317628 | DQ317621 | ||
| PREM 58940 | C. citriodora subsp. variegata | Queensland, Australia | G.S. Pegg | — | — | |||
| Rhodutorula bacarum | CBS 6526T | IGC4391 | Ribes nigrum | United Kingdom | R.W.M. Buhagiar | DQ317629 | AF352055 | |
| R. hinnulea | CBS 8079T | IGC4849 | Banksia collina | Australia | R.G. Shivas | AB038130 | AF190003 | |
| R. phylloplana | CBS 8073T | IGC4246 | B. collina | Australia | R.G. Shivas | DQ317630 | AF190004 | |
| Sympodiomycopsis paphiopedili | CBS 7429T | IGC5543 | nectar of Paphiopedilum primurinum | Japan | K. Tokuoka | DQ317631 | AF190005 | |
| Tilletiopsis pallescens | F3370 | fern leaf | Germany | J.P. Sampaio | DQ317635 | — | ||
| CBS 606.83T | ATCC24345 | basidiome of Sirobasidium sp. | Japan | R.J. Bandoni | DQ317636 | — | ||
| Volvocisporium triumfetticola | R.B. 2070T | Triumfetta rhomboidea | India | M.S. Patil | DQ317637 | AF352053 | ||
Underlined culture collection or herbarium numbers indicate isolates or specimens used in TEM studies.
Holotype specimens or ex-type isolates.
Epitype; CBS = Centraalburaeu voor Schimmelcultures, Utrecht, The Netherlands; CMW = Culture Collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa; R.B. = Herbarium R. Bauer, Tübingen, Germany; F = Culture Collection, Tübingen, Germany; PREM = National Collection of Fungal Specimens, Pretoria, South Africa; IGC = Portugese Yeast Culture Collection, Portugal; ATCC = American Type Culture Collection, Manassas, Virginia, U.S.A.
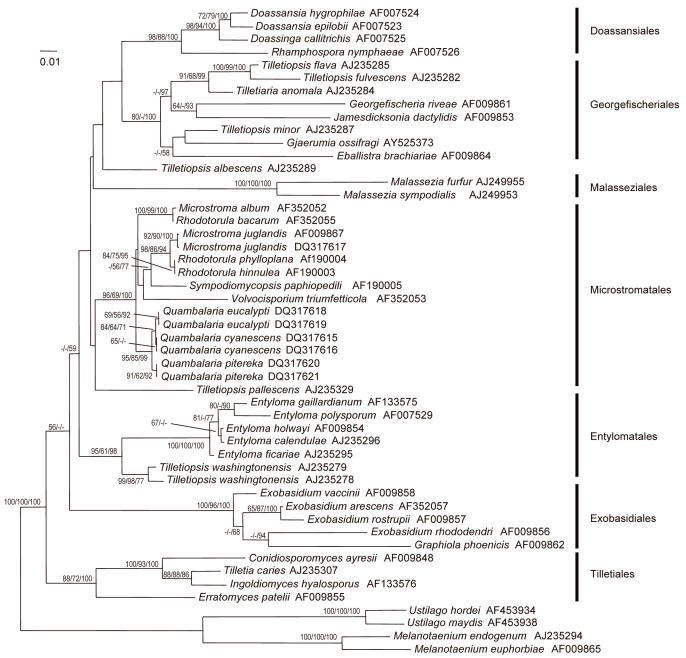
Fig. 1.
Phylogram obtained by neighbour-joining analysis using GTR+I+G substitution model of the nuclear LSU region sequences of species in the Microstromatales. The topology was rooted with four members of the Ustilaginomycetidae. The numbers from left to right refer to percentage bootstrap values of 1000 replicates of neighbour-joining, maximum parsimony, and to a posteriori probabilities of Bayesian Markov chain Monte Carlo analysis. Values smaller than 50 % are not shown. Branch lengths are scaled in terms of expected numbers of nucleotide substitutions per site.
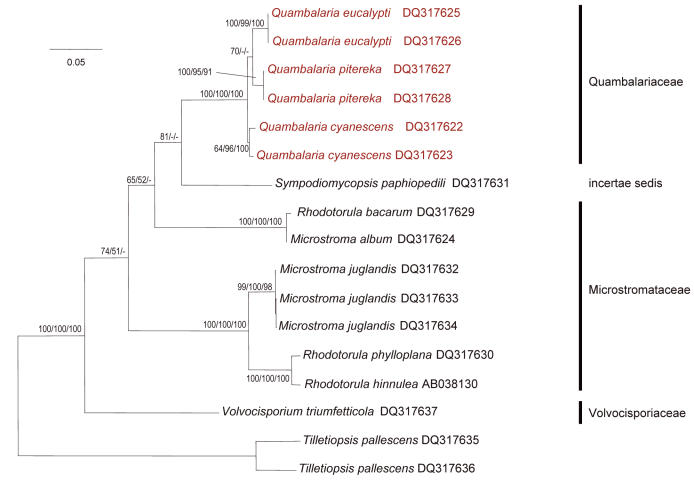
Fig. 2.
Phylogram obtained by neighbour-joining analysis of DNA sequences of the nuclear ITS region of species in the Microstromatales, using the TVM+I+G substitution model. The topology was rooted with two isolates of Tilletiopsis pallescens. The numbers refer to percentage bootstrap values of 1000 replicates of neighbour-joining and maximum parsimony, and to a posteriori probabilities of Bayesian Markov chain Monte Carlo analysis. Values smaller than 50 % are not shown. Branch lengths are scaled in terms of expected numbers of nucleotide substitutions per site.
For ultrastructural examinations of Q. pitereka and Q. eucalypti, herbarium specimens of naturally infected leaves and stems were used (Table 1). These specimens had been deposited in the National Collection of Fungal Specimens, Pretoria, South Africa (PREM). The holotype of Q. eucalypti (PREM 51089) consists of a dried culture on 2 % MEA. However, some important morphological and ultrastructural characters are only expressed on host tissue. The Q. eucalypti specimen we used for ultrastructural work (PREM 58939), consists of symptomatic leaf tissue, collected from the same host in the same location as the holotype (Table 1). This material is designated here as epitype for Q. eucalypti. The culture associated with the epitype (CBS 119680 = CMW 11678), was also included in the phylogenetic analyses. Specimen or isolate numbers of other species in the Microstromatales used for ultrastructural work, are underlined in Table 1.
The ex-type culture of Q. pusilla (U. Braun & Crous) J.A. Simpson (CMW 8279) was found to be contaminated with a Verticillium species and could not be purified. Attempts to extract DNA from the holotype specimen (HAL) were not successful. This species was therefore not included in the study.
DNA extraction and PCR
For the phylogenetic analyses, isolates were grown for 7 d on 2 % malt extract agar. DNA extraction, PCR conditions, visualization and purification of PCR products, as well as DNA sequencing, were done as described by Aghayeva et al. (2004). The internal transcribed spacer region (ITS1, the 5.8S rRNA gene and ITS2), was amplified using PCR with the primers ITS1 and ITS4 (White et al. 1990). The 5' end of the ribosomal large subunit (LSU) was amplified using primers NL1 and NL4 (O'Donnell 1993).
Phylogenetic analyses
Both alignments were assembled with MAFFT 3.85 (Katoh et al. 2002) using the accurate and iterative refinement method (FFT-NS-i settings). After trimming of both ends, the LSU alignment consisted of 572 bp and the ITS alignment of 726 bp. Phylogenetic analyses were carried out using PAUP v. 4.0b10 (Swofford 2001).
Modeltest 3.0 (Posada & Crandall 1998) was applied to determine a model of DNA substitution that fits the data set. GTR+I+G was selected from the Akaike information criterion for the LSU alignment (base frequencies: πA = 0.2563, πC = 0.1950, πG = 0.2911, πT = 0.2576; substitution rates: A/C = 0.7670, A/G = 2.6760, A/T = 0.7823, C/G = 0.3153, C/T = 5.9744, G/T = 1.0000; gamma shape parameter = 0.7950; percentage of invariant sites = 0.3790). TVM+I+G was selected from the Akaike information criterion for the ITS alignment (base frequencies: πA = 0.2535, πC = 0.2188, πG = 0.2157, πT = 0.3120; substitution rates: A/C = 0.14911, A/G C/T = 5.2884, A/T = 2.1848, C/G = 0.8252, G/T = 1.0000; gamma shape parameter = 1.6440; percentage of invariant sites = 0.3892). Neighbour-joining analysis was done determining genetic distances according to the specified substitution model.
Parsimony analysis was conducted in two steps where the first with 10.000 random additions without branch swapping resulted in two islands for the LSU alignment and six for the ITS alignment. Subsequent TBR swapping over the best trees of these islands resulted in four most parsimonious trees for the LSU alignment with 1025 steps (CI = 0.404; RI = 0.665; RC = 0.269), and six trees for the ITS alignment with 507 steps (CI = 0.789; RI = 0.857; RC = 0.676), using 1000 replicates for bootstrap analyses.
For Bayesian analysis, four incrementally heated simultaneous MCMC Markov chains were run over 1 000 000 generations using the general time-reversible model (six rate classes) including a proportion of invariant sites and gamma-distributed substitution rates of the remaining sites (GTR+I+G) (for description of models see Swofford et al. 1996). Trees were sampled every 100th generation, resulting in an overall sampling of 10 000 trees. From these, the first 3000 trees were discarded (as burn-in). MrBayes 3.0b3 (Huelsenbeck & Ronquist 2001) was used to compute a 50 % majority rule consensus of the remaining trees to obtain estimates for the posterior probabilities.
Transmission Electron Microscopy
Species representing the major groups in the Microstromatales, were selected for ultrastructural studies (Table 1). For transmission electron microscopy (TEM), samples were fixed overnight with 2 % glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) at 20 °C. Following six transfers in 0.1M sodium cacodylate buffer, samples were postfixed in 1 % osmium tetroxide in the same buffer for 1 h in the dark, washed in bidistilled water, and stained with 1 % aquaeous uranyl acetate for 1 h in the dark. After five consecutive washes in bidistilled water, samples were dehydrated in acetone, using 10 min transfers at 10, 25, 50, 70, 95, and three times in 100 % acetone. Samples were embedded afterwards in Spurr's plastic and sectioned with a diamond knife. Ultra-thin serial sections were mounted on formvar-coated, single-slot copper grids, stained with lead citrate at room temperature for 5 min, and finally washed with bidistilled water. The samples were studied using a Zeiss EM 109 transmission electron microscope operating at 80 kV.
RESULTS
Phylogenetic analyses
The different phylogenetic analyses of the LSU dataset resulted in similar topologies resolving all known orders of Exobasidiomycetidae Jülich, emend. R. Bauer & Oberw. (Fig. 1). The Tilletiales H. Kreisel ex R. Bauer & Oberw. were weakly supported as sistergroup to the other orders. Although the backbone was not resolved in all parts, the specimens of Quambalaria and Fugomyces considered in this study clustered within the Microstromatales as a highly supported monophylum in both datasets. Tilletiopsis pallescens Gokhale clustered together with members of the Microstromatales and it was, therefore, used as outgroup for the ITS dataset of the Microstromatales.
The ITS regions were used to elucidate the inner phylogeny of the Microstromatales (Fig. 2). Volvocisporium triumfetticola (Patil) Begerow, R. Bauer & Oberw., the only known member of the Volvocisporiaceae Begerow, R. Bauer & Oberw., was sister to the other members of the Microstromatales. Microstroma Niessl appeared paraphyletic in the LSU and ITS analyses, and the relationship between the two Microstroma clusters was weakly supported. This could have resulted from the unclear positions of Sympodiomycopsis paphiopedili Sugiy., Tokuoka & Komag. and V. triumfetticola. All studied specimens of Quambalaria and Fugomyces appeared to form a monophylum. The monophyly of Quambalaria eucalypti and Q. pitereka was supported only in the ITS neighbour-joining analysis and was rejected by maximum parsimony and Bayesian inference and by the LSU data. Quambalaria eucalypti, Q. pitereka and the Fugomyces isolates formed three separate, well-supported clusters. Sequences of the two Q. eucalypti isolates (ex-type and ex-epitype cultures) were identical, and also those of the two Q. pitereka isolates. The ITS sequences of two F. cyanescens isolates differed from each other by 4 bp.
Transmission Electron Microscopy
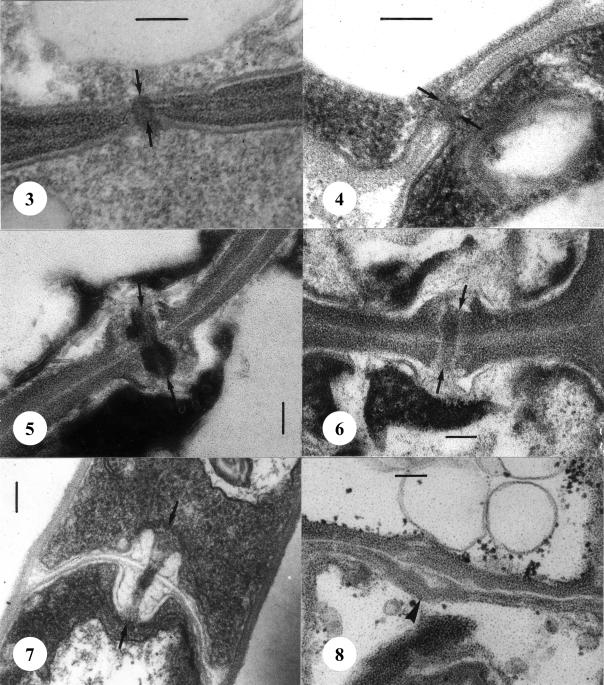
Septal pore apparatuses in the studied species of Microstroma and Volvocisporium Begerow, R. Bauer & Oberw. were simple with more or less rounded pore lips, which were enclosed on both sides by membrane caps (Figs 3–4). In Quambalaria pitereka, Q. eucalypti and Fugomyces cyanescens, the pores were also enclosed by membrane caps, but the septal pore apparatus consisted of dolipores with swollen pore lips (Figs 5–7). In the anamorphic yeast, Sympodiomycopsis paphiopedili we found no septal pores. Occasionally, the septa possess median swellings resembling septal pores, but there was no cytoplasmic continuum between adjacent cells (Fig. 8).
Figs 3–8.
Septation in the Microstromatales. 3. Simple pore with two membrane caps (arrows) of Microstroma juglandis. 4. Simple pore with two membrane caps (arrows) of Volvocisporium triumfetticola. 5. Dolipore of Quambalaria eucalypti with two membrane caps (arrows) from herbarium material. 6. Dolipore with two membrane caps (arrows) of Quambalaria pitereka from herbarium material. 7. Dolipore with two membrane caps (arrows) of Fugomyces cyanescens (CBS 357.73). 8. Pore equivalent in Sympodiomycopsis paphiopedili (CBS 7429). Septum with median swelling (arrowhead), but without cytoplasmic continuim between adjacent cells. Scale bars = 0.1 μm.
TAXONOMY
Phylogenetic analyses of the LSU data obtained in this study showed that the genus Quambalaria resides in the Microstromatales. However, the ultrastructure of the septal pores of Quambalaria spp. differ substantially from those of species in the Microstromataceae Jülich and Volvocisporiaceae. We, therefore, describe a new family, Quambalariaceae, to accommodate the species with dolipores. Thus, the Microstromatales now include not only taxa having septa with simple pores, but also taxa with dolipores or septa without pores. Ultrastructural characteristics, together with LSU and ITS data, show that Fugomyces cyanescens is clearly monophyletic with the two sampled Quambalaria spp. Fugomyces is therefore synonymised here with Quambalaria and the necessary new combination is established.
Quambalariaceae Z.W. de Beer, Begerow & R. Bauer, fam. nov. MycoBank MB500889.
Socii Microstromatalium doliporos cum labiis pororum tumidis facientes.
Members of the Microstromatales having dolipores with swollen pore lips.
Quambalaria J.A. Simpson, Australas. Mycol. 19: 60–61. 2000.
= Fugomyces Sigler, Manual of clinical microbiology, Vol. 2: 1753. 2003.
-
Type species: Quambalaria pitereka (J. Walker & Bertus) J.A. Simpson, Australas. Mycol. 19: 60. 2000.
Basionym: Ramularia pitereka J. Walker & Bertus, Proc. Linn. Soc. New South Wales 96(2): 108. 1971.
- ≡ Sporothrix pitereka (J. Walker & Bertus) U. Braun & Crous, In Braun, A monograph of Ramularia, Cercosporella and allied genera (phytopathogenic hyphomycetes): 416. 1998.
Specimens examined: Australia, Queensland, Corymbia citriodora subsp. variegata leaves, 09 June 1999, M. Ivory, CBS 118828 = CMW 5318; C. citriodora subsp. variegata leaves, 2002, G.S. Pegg, PREM 58940; New South Wales, Grafton, C. maculata leaves, Dec. 2000, M.J. Wingfield, CMW 6707.
-
Quambalaria cyanescens (de Hoog & G.A. de Vries) Z.W. de Beer, Begerow & R. Bauer, comb. nov. MycoBank MB500890.
Basionym: Sporothrix cyanescens de Hoog & G.A. de Vries, Antonie van Leeuwenhoek 39: 515. 1973.
- ≡ Cerinosterus cyanescens (de Hoog & G.A. de Vries) R.T. Moore, Stud. Mycol. 30: 216. 1987.
- ≡ Fugomyces cyanescens (de Hoog & G.A. de Vries) Sigler, In Murray, Manual of clinical microbiology, Vol. 2: 1753. 2003.
Specimens examined: Australia, New South Wales, Armidale, Eucalyptus pauciflora, 1973, V.F. Brown, CBS 876.73 = CMW 5584. Netherlands, Groningen, skin of man, 18 Oct 1959, T.F. Visser, holotype culture ex-type CBS 357.73 = CMW 5583.
-
Quambalaria eucalypti (M.J. Wingf., Crous & W.J. Swart) J.A. Simpson, Australas. Mycol. 19: 61. 2000.
Basionym: Sporothrix eucalypti M.J. Wingf., Crous & W.J. Swart, Mycopathologia 123: 160. 1993.
Specimens examined: South Africa, KwaZulu-Natal, Kwambonambi, Eucalyptus grandis leaves, 19 May 1987, M.J. Wingfield, holotype PREM 51089; KwaZulu-Natal, Kwambonambi, E. grandis leaves, 2001, L. Lombard, PREM 58939, epitype designated here, culture ex-epitype CBS 119680 = CMW 11678.
Species of uncertain status
-
Sporotrichum destructor H.A. Pittman, In Cass Smith, J. Agric. W. Austral. 11 (2): 34. 1970. (nom. nud.)
Note: This fungus, resembling other Quambalaria spp., was isolated by H.A.J. Pittman in 1935 from diseased Corymbia ficifolia in Western Australia. Cultures were sent to Kew where it was identified as a new species named Sporotrichum destructor H.A. Pittman (Cass Smith 1970). However, a Latin diagnosis was never published and material of this species was not available for this study.
-
Quambalaria pusilla (U. Braun & Crous) J.A. Simpson, Australas. Mycol. 19: 61. 2000.
Basionym: Sporothrix pusilla U. Braun & Crous, In Braun, A monograph of Ramularia, Cercosporella and allied genera (phytopathogenic hyphomycetes): 418. 1998.
Note: The ex-type culture of this species (CMW 8279) was contaminated and DNA could not be extracted from the holotype specimen (HAL). The phylogenetic status of this species shall only become clear if fresh material can be obtained.
DISCUSSION
In this study we have produced phylogenetic evidence showing that Q. pitereka infecting Corymbia spp. in Australia and Q. eucalypti, the fungal pathogen on Eucalyptus grandis in South Africa, indeed represent two distinct species. Both LSU and ITS sequence data sets revealed that the two Quambalaria spp. and F. cyanescens (now Q. cyanescens) form a monophyletic lineage in the basidiomycete order Microstromatales. The monophyly of Quambalaria is supported by ultrastructural features. Quambalaria differs from other genera in the Microstromatales because it has dolipores with swollen pore lips in the septa, and not simple pores with more or less rounded pore lips, which are characteristic of the Microstromataceae and Volvocisporiaceae. We have thus described a new family, Quambalariaceae, in the Microstromatales to accommodate Quambalaria spp.
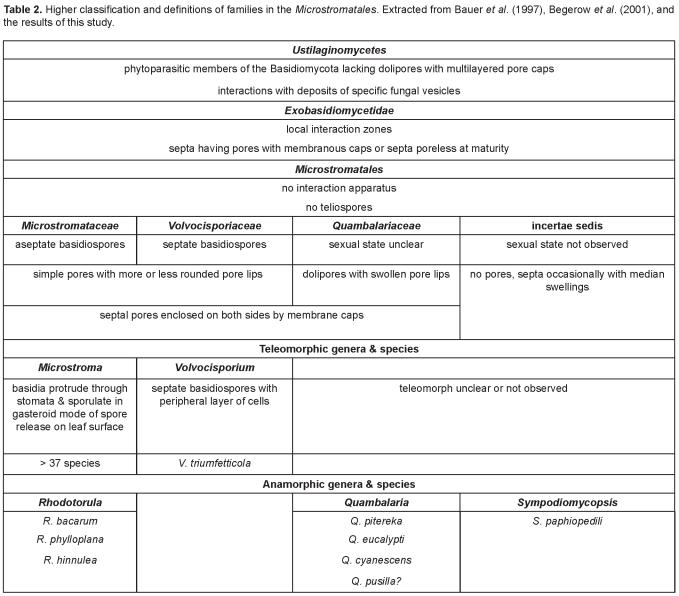
Taxa in the Microstromatales are classified in the subclass Exobasidiomycetidae of the Ustilaginomycetes (Table 2). With few exceptions, the Ustilaginomycetes are restricted to angiosperms, and most are parasites of monocots (Bauer et al. 1997). Of the at least seven orders in the Ustilaginomycetes (Fig. 1), members of only two, the Exobasidiales and the Microstromatales, do not form teliospores and occur on woody bushes or trees (Begerow et al. 2001). The Exobasidiales differ from the Microstromatales by the formation of complex interaction apparatuses including interaction rings (Bauer et al. 1997). The Exobasidiales represent a large order including at least nine genera in four families (Begerow et al. 2002a). The largest of these is Exobasidium Woronin with over 100 species occurring world-wide on flowering plants such as the Ericaceae. Another well-known genus of the Exobasiales is Graphiola Poit., which includes more than 12 species, occurring exclusively on Arecaceae (palms), also with a global distribution (http://nt.arg-grin.gov/fungaldatabases/fungushost/FungusHost.cfm and http://www.indexfungorum.org). A third genus of this order is Muribasidiospora O. Kamat & Rajendren (Begerow et al. 2001). Muribasidiospora indica O. Kamat & Rajendren was recently reported from South Africa for the first time, causing a prominent leaf spot on native Rhus lancea (Crous et al. 2003).
Table 2.
The Microstromatales are characterised by the lack of teliospores and interaction apparatus (Bauer et al. 1997). Only two teleomorphic genera are known in the Microstromatales (Table 2). One of these is Volvocisporium (Table 2 and Fig. 2) which is monotypic. This fungus has such a unique morphology that it was placed in a family of its own (Begerow et al. 2001). The dominant genus in the Microstromatales is Microstroma including about 35 species occurring world-wide, primarily on Leguminosae, Fagaceae and Juglandaceae (http://nt.arg-grin.gov/fungaldatabases/fungushost/FungusHost.cfm and http://www.indexfungorum.org). Only two Microstroma spp. have been reported from South Africa: M. album (Desm.) Sacc. from Quercus, both exotic, and M. albiziae Syd. & P. Syd. from three native Albizia spp. (Doidge 1950). Similarly, two exotic Microstroma spp. have been reported from Australia: again M. album from Quercus and, additionally, M. juglandis (Berenger) Sacc. from Juglans (Sampson & Walker 1982, Shivas 1989). Microstroma album (Fig. 2) is known only from Quercus and has been reported widely from the Northern hemisphere. Microstroma juglandis (Fig. 2) has been found on different genera belonging to the Juglandaceae, with a global distribution. Microstroma albiziae has only been reported from Albizia spp. in South Africa (Doidge 1950) and India (Mathur 1979). Material of these species was not available for study.
Begerow et al. (2001) showed with LSU sequence analyses that two anamorphic yeasts, Rhodotorula bacarum (Buhagiar) Rodr. Mir. & Weijman and R. phylloplana (R.G. Shivas & Rodr. Mir.) Rodr. Mir. & Weijman are phylogenetically closely related to Microstroma album and M. juglandis, respectively. Our ITS data (Fig. 2), support their results and show that R. bacarum might be the same species as M. album. We included a third species, R. hinnulea (R.G. Shivas & Rodr. Mir.) Rodr. Mir. & Weijman, and it differs from R. phylloplana in only 2 bp. (Fig. 2). Both these species were isolated from the leaves of Banksia collina (Proteaceae) in Australia, and were described then as new Cryptococcus species (Shivas & Rodrigues de Miranda 1983). However, the biochemical and morphological differences (Shivas & Rodrigues de Miranda 1983) between the two species are small and they might represent individuals of the same species. The three Rhodotorula spp. should not be accommodated in the genus Rhodotorula, because the type species for Rhodotorula, R. glutinis (Fresen.) F.C. Harrison, is phylogenetically (based on sequence data) placed in the Sporidiales R.T. Moore in the Urediniomycetes (Swann & Taylor 1995). We have chosen not to erect a new anamorph genus for these fungi at the present time, since they might be linked to teleomorphs (probably Microstroma spp.) and could be more appropriately treated at a time when additional material is available for study.
The monophyly (Fig. 2) and ultrastructural similarities (Figs 5–7) between the three Quambalaria spp. recognised in this study, is supported by the ecology of these species. The fact that all three species, as well as Q. pusilla (not included), occur on tree species native to Australia, suggests that Australia is the centre of origin of these species. Although Q. cyanescens has been isolated from human tissues on several occasions, the fungus has not been associated with specific disease symptoms of humans (Middelhoven et al. 2000, Sigler & Verweij 2003). Inoculation trials on mice failed to demonstrate virulence of the fungus on mammals (Sigler et al. 1990). The fungus is, therefore, rather regarded as an opportunist, and potentially can be implicated in disease in immunocompromised patients (Tambini et al. 1996).
The recognition of Quambalaria spp. as basidiomycetes has not been widely considered because the teleomorph has never been observed. When the teleomorph morphology of the closely related fungus M. juglandis is considered (Begerow et al. 2001), it might be found that the teleomorph of Quambalaria is masquerading as an anamorph. This is entirely possible as the anamorph and teleomorph states would be difficult to distinguish from each other.
One of the species for which the position in the Microstromatales remains uncertain (Table 2 and Fig. 2), is the anamorphic yeast Sympodiomycopsis paphiopedili. This fungus was described from the nectar of an orchid in Japan (Sugiyama et al. 1991). Although the conidiogenous cells in culture (Sugiyama et al. 1991) resemble those of Quambalaria, its phylogenetic position (Fig. 2) sets it apart from all the other members of the Microstromatales. Because this yeast forms pseudomycelia, occasionally with retraction septa, it is not surprising that we did not observe pores (Bauer et al. 2001), but septa with median swellings (Fig. 8). Suh et al. (1993) reported simple pores in S. paphiopedili, but the respective micrograph is insufficient. The pore structure of the hyphal phase of S. paphiopedili is thus unknown.
Recognition of three families in the Microstromatales and emerging lineages that correspond with host families, follows a trend that has been observed in other orders in the Ustilaginomycetes (Begerow et al. 2004). The four families in the Exobasidiales, for example, can be distinguished based on basidial morphology and host range, but these characteristics also match phylogenetic lineages based on LSU rDNA sequences (Begerow et al. 2002a). Cospeciation of groups of species in the Entylomatales R. Bauer & Oberw. with their hosts, has also been shown (Begerow et al. 2002b). To test cospeciation processes in the Microstromatales, additional fungal isolates from a wider variety of hosts would need to be included in phylogenetic studies together with their host species. However, there is good evidence that Q. pitereka infects only Corymbia and Q. eucalypti is restricted to hosts in the genus Eucalyptus. These two tree genera are phylogenetically distinct (Hill & Johnson 1995, Wilson et al. 2001) and it appears that the pathogens have specifically evolved to infect them.
Studies on members of the Microstromatales have been limited, most likely because they have not been considered an economically important group of fungi. This perception is changing rapidly with the reported spread of disease caused by members of the Quambalariaceae in commercial Eucalyptus plantations in South Africa (Wingfield et al. 1993), Brazil and Uruguay (Alfenas et al. 2001, Zauza et al. 2003), and in Corymbia plantations in Australia (Simpson 2000, Pegg et al. 2005). That we have only touched the “tip of the iceberg” of the Microstromatales (Begerow et al. 2001) should be regarded as a challenge, since so many questions surrounding the biology and distribution of this intriguing group of fungi remain unanswered.
Acknowledgments
We thank Dr Hugh Glen for the Latin diagnosis and Dr XuDong Zhou for assistance with some of the sequences in the laboratory. We acknowledge the members of the Tree Protection Co-operative Programme (TPCP), the department of Trade and Industry (DTI) THRIP initiative, the National Research Foundation (NRF), the NRF/DST Centre of Excellence in Tree Health Biotechnology (CTHB), and the Deutsche Forschungsgemeinschaft (DFG) for financial support.
Taxonomic novelties: Quambalariaceae Z.W. de Beer, Begerow & R. Bauer fam. nov., Quambalaria cyanescens (de Hoog & G.A. de Vries) Z.W. de Beer, Begerow & R. Bauer comb. nov.
References
- Aghayeva DN, Wingfield MJ, De Beer ZW, Kirisits T (2004). Two new Ophiostoma species with Sporothrix anamorphs from Austria and Azerbaijan. Mycologia 96: 866–878. [DOI] [PubMed] [Google Scholar]
- Alfenas AC, Zauza EAV, Rosa OPP, Assis TF (2001). Sporothrix eucalypti, um novo patógeno do eucalipto no Brazil. Fitopatologia Brasileira 26: 221. [Google Scholar]
- Bauer R, Begerow D, Oberwinkler F, Piepenbring M, Berbee ML (2001). Ustilaginomycetes. In: Mycota VII, Part B. Systematics and evolution (McLaughlin DJ, McLaughlin EG, Lemke PA, eds). Springer Verlag, Heidelberg: 57–83.
- Bauer R, Oberwinkler F, Vánky K (1997). Ultrastructural markers and systematics in smut fungi and allied taxa. Canadian Journal of Botany 75: 1273–1314. [Google Scholar]
- Begerow D, Bauer R, Oberwinkler F (1997). Phylogenetic studies on nuclear large subunit ribosomal DNA sequences of smut fungi and related taxa. Canadian Journal of Botany 75: 2045–2056. [Google Scholar]
- Begerow D, Bauer R, Oberwinkler F (2001). Muribasidiospora: Microstromatales or Exobasidiales? Mycological Research 105: 798–810. [Google Scholar]
- Begerow D, Bauer R, Oberwinkler F (2002a). The Exobasidiales: an evolutionary hypothesis. Mycological Progress 1: 187–199. [Google Scholar]
- Begerow D, Göker M, Lutz M, Stoll M (2004). On the evolution of smuts and their hosts. In: Frontiers in Basidiomycete Mycology (Agerer, Piepenbring, Blanz, eds). IHW-Verlag, Eching: 81–98.
- Begerow D, Lutz M, Oberwinkler F (2002b). Implications of molecular characters for the phylogeny of the genus Entyloma. Mycological Research 106: 1392–1399. [Google Scholar]
- Berbee ML, Taylor JW (1992). 18S Ribosomal RNA gene sequence characters place the human pathogen Sporothrix schenckii in the genus Ophiostoma. Experimental Mycology 16: 87–91. [Google Scholar]
- Braun U (1998). A monograph of Ramularia, Cercosporella and allied genera (phytopathogenic hyphomycetes). IHW-Verlag, Eching.
- Cass Smith WP (1970). Stem canker disease of red flowering gums. Journal of the Department of Agriculture, Western Australia 11: 33–39. [Google Scholar]
- Crous PW, Groenewald JZ, Carroll G (2003). Muribasidiospora indica causing a prominent leaf spot disease on Rhus lancea in South Africa. Australasian Plant Pathology 32: 313–316. [Google Scholar]
- De Hoog GS (1993). Sporothrix-like anamorphs of Ophiostoma species and other fungi. In: Ceratocystis and Ophiostoma: Taxonomy, Ecology and Pathogenicity (Wingfield MJ, Seifert KA, Webber J, eds). American Phytopathological Society, St. Paul, Minnesota: 53–60.
- De Hoog GS, De Vries GA (1973). Two new species of Sporothrix and their relation to Blastobotrys nivea. Antonie van Leeuwenhoek 39: 515–520. [DOI] [PubMed] [Google Scholar]
- Doidge EM (1950). The South African fungi and lichens to the end of 1945. Bothalia 5: 1–1094. [Google Scholar]
- Harrington TC (1981). Cycloheximide sensitivity as a taxonomic character in Ceratocystis. Mycologia 73: 1123–1129. [Google Scholar]
- Hill KD, Johnson LAS (1995). Systematic studies in the eucalypts – 7. A revision of the bloodwoods, genus Corymbia (Myrtaceae). Telopea 6: 173–505. [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur RS (1979). The Coelomycetes of India. Bishen Singh Mahendra Pal Singh, Delhi, India.
- Middelhoven WJ, Guého E, De Hoog GS (2000). Phylogenetic position and physiology of Cerinosterus cyanescens. Antonie van Leeuwenhoek 77: 313–320. [DOI] [PubMed] [Google Scholar]
- Moore RT (1987). Micromorphology of yeasts and yeast-like fungi and its taxonomic implications. Studies in Mycology 30: 203–226. [Google Scholar]
- Pegg GS, Drenth A, Wingfield MJ (2005). Quambalaria pitereka on spotted gum plantations in Queensland and northern New South Wales, Australia. Abstracts of XII IUFRO World Congress, 8–13 August 2005, Brisbane, Australia. International Forestry Review 7: 337. [Google Scholar]
- Posada D, Crandall KA (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Sampson PJ, Walker J (1982). An Annotated List of Plant Diseases in Tasmania. Department of Agriculture Tasmania.
- Shivas RG (1989). Fungal and bacterial diseases of plants in Western Australia. Journal of the Royal Society of Western Australia 72: 1–62. [Google Scholar]
- Shivas RG, Rodrigues de Miranda L (1983). Cryptococcus phylloplanus and Cryptococcus hinnuleus, two new yeast species. Antonie van Leeuwenhoek 49: 153–158. [DOI] [PubMed] [Google Scholar]
- Sigler L, Harris JL, Dixon DM (1990). Microbiology and potential virulence of Sporothrix cyanescens, a fungus rarely isolated from blood and skin. Journal of Clinical Microbiology 29: 1009–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler L, Verweij PE (2003). Aspergillus, Fusarium, and other opportunistic moniliaceous fungi. In: Manual of Clinical Microbiology (Murray PR, ed). ASM Press, Washington, D.C.: 1726–1760.
- Simpson JA (2000). Quambalaria, a new genus of eucalypt pathogens. Australasian Mycologist 19: 57–62. [Google Scholar]
- Smith MT, Batenburg–Van der Vegte WH (1985). Ultrastructure of septa in Blastobotrys and Sporothrix. Antonie van Leeuwenhoek 51: 121–128. [DOI] [PubMed] [Google Scholar]
- Sugiyama J, Tokuoka K, Suh S–O, Hirata A, Komagata K (1991). Sympodiomycopsis: a new yeast-like anamorph genus with basidiomycetous nature from orchid nectar. Antonie van Leeuwenhoek 59: 95–108. [DOI] [PubMed] [Google Scholar]
- Suh S–O, Hirata A, Sugiyama J, Komagata K (1993). Septal ultrastructure of basidiomycetous yeasts and their taxonomic implications with observations on the ultrastructure of Erythrobasidium hasegawianum and Sympodiomycopsis paphiopedili. Mycologia 85: 30–37. [Google Scholar]
- Suzuki M, Nakase T (1986). Heterogeneity of ubiquinone systems in the genus Sporothrix. Journal of General and Applied Microbiology 32: 165–168. [Google Scholar]
- Swann EC, Taylor JW (1995). Phylogenetic diversity of yeast-producing basidiomycetes. Mycological Research 99: 1205–1210. [Google Scholar]
- Swofford DL (2001). PAUP*. Phylogenetic analysis using parsimony (*and other methods) version 4.0b1. Sinauer Associates, Sunderland, Massachusetts.
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM (1996). Phylogenetic Inference. In: Molecular Systematics (Hillis DM, Moritz C, Mable BK, eds). Sinauer Associates Inc., Sunderland, Massachusetts, U.S.A.: 407–514.
- Tambini R, Farina C, Fiocchi R, Dupont B, Guého E, Delvecchio G, Mamprin F, Gavazzeni G (1996). Possible pathogenic role for Sporothrix cyanescens isolated from a lung lesion in a heart transplant patient. Medical Mycology 34: 195–198. [PubMed] [Google Scholar]
- Walker J, Bertus AL (1971). Shoot blight of Eucalyptus spp. caused by an undescribed species of Ramularia. Proceedings of the Linnean Society of New South Wales 96: 108–115. [Google Scholar]
- Weijman ACM, De Hoog GS (1985). Carbohydrate patterns and taxonomy of Sporothrix and Blastobotrys. Antonie van Leeuwenhoek 51: 111–120. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: A guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds). Academic Press, Inc., San Diego, California: 315–322.
- Wilson PG, O'Brien MM, Gadek PA, Quinn CJ (2001). Myrtaceae revisited: a reassessment of infrafamilial groups. American Journal of Botany 88: 2013–2025. [PubMed] [Google Scholar]
- Wingfield MJ, Crous PW, Swart WJ (1993). Sporothrix eucalypti (sp. nov.), a shoot and leaf pathogen of Eucalyptus in South Africa. Mycopathologia 123: 159–164. [Google Scholar]
- Zauza EAV, Alfenas AC, Langrell SRH, Tommerup IC (2003). Detection and identification of Quambalaria species in Eucalyptus nurseries and plantations. 8th International Congress of Plant Pathology, Christchurch, New Zealand.