Abstract
Saprobic Cladosporium isolates morphologically similar to C. sphaerospermum are phylogenetically analysed on the basis of DNA sequences of the ribosomal RNA gene cluster, including the internal transcribed spacer regions ITS1 and ITS2, the 5.8S rDNA (ITS) and the small subunit (SSU) rDNA as well as β-tubulin and actin gene introns and exons. Most of the C. sphaerospermum-like species show halotolerance as a recurrent feature. Cladosporium sphaerospermum, which is characterised by almost globose conidia, is redefined on the basis of its ex-neotype culture. Cladosporium dominicanum, C. psychrotolerans, C. velox, C. spinulosum and C. halotolerans, all with globoid conidia, are newly described on the basis of phylogenetic analyses and cryptic morphological and physiological characters. Cladosporium halotolerans was isolated from hypersaline water and bathrooms and detected once on dolphin skin. Cladosporium dominicanum and C. velox were isolated from plant material and hypersaline water. Cladosporium psychrotolerans, which grows well at 4 °C but not at 30 °C, and C. spinulosum, having conspicuously ornamented conidia with long digitate projections, are currently only known from hypersaline water. We also newly describe C. salinae from hypersaline water and C. fusiforme from hypersaline water and animal feed. Both species have ovoid to ellipsoid conidia and are therefore reminiscent of C. herbarum. Cladosporium langeronii (= Hormodendrum langeronii) previously described as a pathogen on human skin, is halotolerant but has not yet been recorded from hypersaline environments.
Keywords: Actin, β-tubulin, halotolerance, ITS rDNA, phylogeny, SSU rDNA, taxonomy
INTRODUCTION
The halophilic and halotolerant mycobiota from hypersaline aqueous habitats worldwide frequently contain Cladosporium Link isolates (Gunde-Cimerman et al. 2000, Butinar et al. 2005). Initially, they were considered as airborne contaminants, but surprisingly, many of these Cladosporium isolates were identified as C. sphaerospermum Penz. because they formed globoid conidia (data unpublished). Cladosporium sphaerospermum, known as one of the most common air-borne, cosmopolitan Cladosporium species, was frequently isolated from indoor and outdoor air (Park et al. 2004), dwellings (Aihara et al. 2001), and occasionally from humans (Badillet et al. 1982) and plants (Pereira et al. 2002). Strains morphologically identified as C. sphaerospermum were able to grow at a very low water activity (aw 0.816), while other cladosporia clearly preferred a higher, less extreme water activity (Hocking et al. 1994). This pronounced osmotolerance suggests a predilection for osmotically stressed environments although C. sphaerospermum is reported from a wide range of habitats including osmotically non-stressed niches.
We therefore hypothesised that C. sphaerospermum represents a complex of species having either narrow or wide ecological amplitudes. The molecular diversity of strains identified as C. sphaerospermum has not yet been determined and isolates from humans have not yet been critically compared with those from environmental samples. Therefore, a taxonomic study was initiated with the aim to define phylogenetically and morphologically distinct entities and to describe their in vitro osmotolerance and their natural ecological preferences.
MATERIALS AND METHODS
Sampling
Samples of hypersaline water were collected from salterns located at different sites of the Mediterranean basin (Slovenia, Bosnia and Herzegovina, Spain), different coastal areas along the Atlantic Ocean (Monte Cristy, Dominican Republic; Swakopmund, Namibia), the Red Sea (Eilat, Israel), the Dead Sea (Ein Gedi, Israel), and the salt Lake Enriquillio (Dominican Republic). Samples from the Sečovlje salterns (Slovenia) were collected once per month in 1999. Samples from the Santa Pola salterns and Ebre delta river saltern (Spain) were taken twice (July and November) in 2000. A saltern in Namibia and one in the Dominican Republic were sampled twice (August and October) in 2002. Various salinities, ranging from 15 to 32 % NaCl were encountered in these ponds.
Isolation and maintenance of fungi
Strains were isolated from salterns using filtration of hypersaline water through membrane filters (pore diam 0.45 μm), followed by incubation of the membrane filters on different culture media with lowered water activity (Gunde-Cimerman et al. 2000). Only colonies of different morphology on one particular selective medium per sample were analysed further. Strains were carefully selected from different evaporation ponds, collected at different times, in order to avoid sampling of identical clones. Subcultures were maintained at the Culture Collection of Extremophilic Fungi (EXF, Biotechnical Faculty, Ljubljana, Slovenia), while a selection was deposited at the Centraalbureau voor Schimmelcultures (CBS, Utrecht, The Netherlands) and the Culture Collection of the National Institute of Chemistry (MZKI, Ljubljana, Slovenia). Reference strains were obtained from CBS, and were selected either on the basis of the strain history, name, or on the basis of their ITS rDNA sequence. Strains were maintained on oatmeal agar (OA; diluted OA, Difco: 15 g of Difco 255210 OA medium, 12 g of agar, dissolved in 1 L of distilled water) with or without 5 % additional NaCl. They were preserved in liquid nitrogen or by lyophilisation. Strains studied are listed in Table 1.
Table 1.
List of Cladosporium strains, with their current and original names, geography, GenBank accession numbers and references to earlier published sequences.
| Strain Nr.a | Source | Geography |
GenBank accession
Nr.b
|
|
|
|---|---|---|---|---|---|
| ITS rDNA / 18S rDNA | actin | β-tubulin | |||
| Cladosporium bruhnei | |||||
| CBS 177.71 | Thuja tincture | The Netherlands, Amsterdam | DQ780399 / DQ780938 | EF101354 | EF101451 |
| CBS 812.71 | Polygonatum odoratum, leaf | Czech Republic, Lisen | DQ780401 / - | - | - |
| Cladosporium cladosporioides | |||||
| CBS 170.54NT | Arundo, leaf | U.K., England, Kew | AY213640 / DQ780940 | EF101352 | EF101453 |
| EXF-321 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780408 / - | - | - |
| EXF-780 | DQ780409 / - | - | - | ||
| EXF-946 | Hypersaline water | Bosnia and Herzegovina, Ston saltern | DQ780410 / - | - | - |
| Cladosporium dominicanum | |||||
| CPC 11683 | Citrus fruit (orange) | Iran | DQ780357 / - | EF101369 | EF101419 |
| EXF-696 | Hypersaline water | Dominican Republic, saltern | DQ780358 / - | EF101367 | EF101420 |
| EXF-718 | Hypersaline water | Dominican Republic, salt lake Enriquilio | DQ780356 / - | EF101370 | EF101418 |
| EXF-720 | Hypersaline water | Dominican Republic, saltern | DQ780355 / - | - | EF101417 |
| EXF-727 | Hypersaline water | Dominican Republic, saltern | DQ780354 / - | - | EF101416 |
| EXF-732 T; CBS 119415 | Hypersaline water | Dominican Republic, salt lake Enriquilio | DQ780353 / - | EF101368 | EF101415 |
| Cladosporium fusiforme | |||||
| CBS 452.71 | Chicken food | Canada | DQ780390 / - | EF101371 | EF101447 |
| EXF-397 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780389 / - | EF101373 | EF101445 |
| EXF-449 T; CBS 119414 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780388 / DQ780935 | EF101372 | EF101446 |
| Cladosporium herbarum | |||||
| ATCC 66670, as Davidiella tassiana | CCA-treated Douglas-fir pole | U.S.A., New York, Geneva | AY3619592 & DQ780400 / DQ780939 | AY75219311 | EF101452 |
| Cladosporium halotolerans | |||||
| ATCC 26362 | Liver and intestine of diseased frog | U.S.A., New Jersey | AY3619822 / - | - | - |
| ATCC 64726 | Peanut cell suspension tissue culture | U.S.A., Georgia | AY3619682 / - | - | - |
| CBS 280.49 | Stem of Hypericum perforatum identified as Mycosphaerella hyperici | Switzerland, Glarus, Mühlehorn | DQ780369 / - | EF101402 | EF101432 |
| CBS 191.54 | Laboratory air | Great Britain | - / - | - | - |
| CBS 573.78 | Aureobasidium caulivorum | Russia, Moscow region | - / - | - | - |
| CBS 626.82 | - | Sweden, Stockholm | - / - | - | - |
| dH 12862; EXF-2533 | Culture contaminant | Brazil | DQ780371 / - | EF101400 | EF101422 |
| dH 12941; EXF-2534 | Culture contaminant | Turkey | - / - | EF101421 | |
| dH 12991; EXF-2535 | Brain | Turkey | DQ780372 / - | EF101423 | |
| dH 13911; EXF-2422 | Ice | Arctics | DQ780370 / - | EF101401 | EF101430 |
| EXF-228; MZKI B-840 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780365 / DQ780930 | EF101393 | EF101425 |
| EXF-380 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780368 / - | EF101394 | EF101427 |
| EXF-564 | Hypersaline water | Namibia, saltern | DQ780363 / - | EF101395 | EF101433 |
| EXF-565 | Hypersaline water | Namibia, saltern | - / - | - | - |
| EXF-567 | Hypersaline water | Namibia, saltern | - / - | - | - |
| EXF-571 | Hypersaline water | Namibia, saltern | - / - | - | - |
| EXF-572 T; CBS 119416 | Hypersaline water | Namibia, saltern | DQ780364 / - | EF101397 | EF101424 |
| EXF-646 | Hypersaline water | Spain, Santa Pola saltern | DQ780366 / - | EF101398 | EF101428 |
| EXF-698 | Hypersaline water | Dominican Republic, saltern | - / - | - | - |
| EXF-703 | Hypersaline water | Dominican Republic, salt lake Enriquilio | DQ780367 / - | EF101392 | EF101426 |
| EXF-944 | Hypersaline water | Bosnia and Herzegovina, Ston saltern | - / - | - | - |
| EXF-972 | Bathroom | Slovenia | - / - | - | - |
| EXF-977 | Bathroom | Slovenia | DQ780362 / - | EF101396 | EF101431 |
| EXF-1072 | Hypersaline water | Israel, Dead Sea | DQ780373 / - | EF101399 | EF101428 |
| EXF-2372 | Hypersaline water | Slovenia, Sečovlje saltern | - / - | - | - |
| UAMH 7686 | Indoor air ex RCS strip, from Apis mellifera overwintering facility | U.S.A., Alta, Clyde Corner | AY6250635 / - | - | - |
| - | rDNA from bottlenose dolphin skin infected with Loboa loboi | U.S.A., Texas | AF0356746 / - | - | - |
| - | Microcolony, on rock | Turkey, Antalya | AJ9714097 / - | - | - |
| - | Microcolony, on rock | Turkey, Antalya | AJ9714087 / - | - | - |
| - | Tomato leaves | - | L254338 / - | - | - |
| Cladosporium langeronii | |||||
| CBS 189.54NT | Mycosis | Brazil | DQ780379 / DQ780932 | EF101357 | EF101435 |
| CBS 601.84 | Picea abies, wood | Germany, Göttingen | DQ780382 / - | EF101360 | EF101438 |
| CBS 101880 | Moist aluminium school window frame | Belgium, Lichtervoorde | DQ780380 / - | EF101359 | EF101440 |
| CBS 109868 | Mortar of Muro Farnesiano | Italy, Parma | DQ780377 / - | EF101362 | EF101434 |
| dH 11736 | Biomat in a lake | Antarctics | DQ780381 / - | EF101363 | EF101436 |
| dH 12459 | Orig. face lesion | Brazil | DQ780378 / - | EF101358 | EF101439 |
| dH 13833 | Ice | Arctics | DQ780383 / - | EF101361 | EF101437 |
| - | Nasal mucus | - | AF4555254 / - | - | - |
| - | Nasal mucus | - | AY3453524 / - | - | - |
| - | Mycorrhizal roots | - | DQ0689829 / - | - | - |
| Cladosporium oxysporum | |||||
| ATCC 66669 | Creosote-treated southern pine pole | U.S.A., New York, Binghamton | AF39368910 / DQ780395 | AY75219211 | EF101454 |
| ATCC 76499 | Decayed leaf, Lespedeza bicolor | - | AF393720 | - | - |
| CBS 125.80 | Cirsium vulgare, seedcoat | The Netherlands | AJ30033212 / DQ780941 | EF101351 | EF101455 |
| EXF-697 | Hypersaline water | Dominican Republic, salt lake Enriquilio | DQ780392 / - | - | - |
| EXF-699 | Hypersaline water | Dominican Republic, saltern | DQ780394 / - | - | - |
| EXF-710 | Hypersaline water | Dominican Republic, saltern | DQ780393 / - | - | - |
| EXF-711 | Hypersaline water | Dominican Republic, saltern | DQ780391 / - | - | - |
| Cladosporium psychrotolerans | |||||
| EXF-326 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780387 / DQ780934 | - | EF101444 |
| EXF-332 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780385 / DQ780933 | EF101364 | EF101441 |
| EXF-391 T; CBS 119412 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780386 / - | EF101365 | EF101442 |
| EXF-714 | Hypersaline water | Dominican Republic | DQ780384 / - | EF101366 | EF101443 |
| Cladosporium ramotenellum | |||||
| EXF-454 T; CPC 12043 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780403 / - | - | - |
| Cladosporium salinae | |||||
| EXF-322 | Hypersaline water | Slovenia, Sečovlje | DQ780375 / - | EF101391 | EF101403 |
| EXF-335 T; CBS 119413 | Hypersaline water | Slovenia, Sečovlje | DQ780374 / DQ780931 | EF101390 | EF101405 |
| EXF-604 | Hypersaline water | Spain, Santa Pola | DQ780376 / - | EF101389 | EF101404 |
| Cladosporium sp. | |||||
| CBS 300.96 | Soil along coral reef coast | Papua New Guinea, Madang, Jais Aben | DQ780352 / - | EF101385 | - |
| EXF-595 | Hypersaline water | Spain, Santa Pola saltern | DQ780402 / - | - | - |
| Cladosporium sphaerospermum | |||||
| ATCC 12092 | Soil | Canada | AY3619882 / - | - | - |
| ATCC 200384 | Compost biofilter | The Netherlands | AY3619912 / - | - | - |
| CBS 109.14; ATCC 36950 | Carya illinoensis leaf scale | U.S.A. | DQ780350 / - | EF101384 | EF101410 |
| CBS 122.47; IFO 6377; IMI 49640; VKM F-772; ATCC 11292 | Decaying stem of Begonia sp., with Thielaviopsis basicola | The Netherlands, Aalsmeer | AJ2442281 / - | - | - |
| CBS 188.54; ATCC 11290; IMI 049638 | de Vries (Engelhardt strain) | - | AY3619902 & AY2510773 / - | - | - |
| CBS 190.54; ATCC 11293; IFO 6380; IMI 49641 | - | - | AY3619922 / - | - | - |
| CBS 192.54; ATCC 11288; IMI 49636 | Nail of man | - | AY3619892 / - | - | - |
| CBS 193.54NT; ATCC 11289; IMI 49637 | Human nails | - | DQ780343 & AY3619582 / DQ780925 | EF101380 | EF101406 |
| CBS 122.63 | Plywood of Betula sp. | Finland, Helsinki | - / - | - | - |
| CBS 102045; EXF-2524; MZKI B-1066 | Hypersaline water | Spain, Barcelona, Salines de la Trinitat | DQ780351 / - | EF101378 | EF101411 |
| CBS 114065 | Outdoor air | Germany, Stuttgart | - / - | - | - |
| CPC 10944 | Gardening peat substrate | Russia, Kaliningrad | DQ780350 / - | - | - |
| EXF-131; MZKI B-1005 | Hypersaline water | Slovenia, Sečovlje saltern | AJ2386701 / - | - | - |
| EXF-328 | Hypersaline water | Slovenia, Sečovlje saltern | - / - | - | - |
| EXF-385 | Hypersaline water | Slovenia, Sečovlje saltern | - / - | - | - |
| EXF-446 | Hypersaline water | Slovenia, Sečovlje saltern | - / - | - | - |
| EXF-455 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780349 / - | EF101375 | EF101412 |
| EXF-458 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780345 / - | EF101374 | EF101409 |
| EXF-461 | Hypersaline water | Slovenia, Sečovlje saltern | - / - | - | - |
| EXF-464 | Hypersaline water | Slovenia, Sečovlje saltern | - / DQ780927 | - | - |
| EXF-465 | Hypersaline water | Slovenia, Sečovlje saltern | - / - | - | - |
| EXF-598 | Hypersaline water | Spain, Santa Pola | - / - | EF101377 | - |
| EXF-644 | Hypersaline water | Spain, Santa Pola | - / - | - | - |
| EXF-645 | Hypersaline water | Spain, Santa Pola | - / - | - | - |
| EXF-649 | Hypersaline water | Spain, Santa Pola | - / - | - | - |
| EXF-715 | Hypersaline water | Dominican Republic, saltern | - / - | - | - |
| EXF-738 | Bathroom | Slovenia | DQ780348 / - | EF101383 | EF101414 |
| EXF-739 | Bathroom | Slovenia | DQ780344 / - | EF101381 | EF101407 |
| EXF-781; MZKI B-899 | Hypersaline water | Slovenia, Sečovlje | - / - | - | - |
| EXF-788 | Hypersaline water | Slovenia, Sečovlje | - / - | - | - |
| EXF-962 | Bathroom | Slovenia | DQ780347 / - | EF101382 | EF101413 |
| EXF-965 | Bathroom | Slovenia | - / - | - | - |
| EXF-1069 | Hypersaline water | Israel, Eilat saltern | - / - | EF101376 | - |
| EXF-1061 | Hypersaline water | Israel, Dead Sea | DQ780346 / - | EF101379 | EF101408 |
| EXF-1726 | Hypersaline water | Israel, Dead Sea | - / - | - | - |
| EXF-1732 | Hypersaline water | Israel, Eilat saltern | - / DQ780928 | - | - |
| - | Bryozoa sp. | - | AJ557744 / - | - | - |
| - | Nasal mucus | - | AF4554814 / - | - | - |
| Cladosporium spinulosum | |||||
| EXF-333 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780404 / - | - | - |
| EXF-334 T | Hypersaline water | Slovenia, Sečovlje saltern | DQ780406 / - | EF101355 | EF101450 |
| EXF-382 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780407 / DQ780936 | EF101356 | EF101449 |
| Cladosporium subinflatum | |||||
| EXF-343 T; CPC 12041 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780405 / - | EF101353 | EF101448 |
| Cladosporium tenuissimum | |||||
| ATCC 38027 | Soil | New Caledonia | AF393724 / - | - | - |
| EXF-324 | Hypersaline water | Slovenia, Sečovlje saltern | - / DQ780926 | - | - |
| EXF-371 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780396 / - | - | - |
| EXF-452 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780397 / - | - | - |
| EXF-563 | Hypersaline water | Namibia, saltern | DQ780398 / - | - | - |
| Cladosporium velox | |||||
| CBS 119417T; CPC 11224 | Bamboo sp. | India, Charidij | DQ780361 / DQ780937 | EF101388 | EF101456 |
| EXF-466 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780359 / - | EF101386 | - |
| EXF-471 | Hypersaline water | Slovenia, Sečovlje saltern | DQ780360 / - | EF101387 | - |
Abbreviations used: ATCC = American Type Culture Collection, Virginia, U.S.A.; CBS = Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CPC = Culture Collection of Pedro Crous, housed at CBS, Utrecht, The Netherlands; dH = de Hoog Culture Collection, housed at CBS, Utrecht, The Netherlands; EXF = Culture Collection of Extremophilic Fungi, Ljubljana, Slovenia; IFO = Institute for Fermentation, Culture Collection of Microorganizms, Osaka, Japan; IMI = The International Mycological Institute, Egham, Surrey, U.K.; MZKI = Microbiological Culture Collection of the National Institute of Chemistry, Ljubljana, Slovenia; UAMH = University of Alberta Microfungus Collection, Alberta, Canada; VKM = All-Russian Collection of Microorganisms, Russian Academy of Sciences, Institute of Biochemistry and Physiology of Microorganisms, Pushchino, Russia; NT = ex-neotype strain; T = ex-type strain.
Reference: 1de Hoog et al. 1999; 2Park et al. 2004; 3Braun et al. 2003; 4Buzina et al. 2003; 5Meklin et al. 2004; 6Haubold et al. 1998; 7Sert & Sterflinger, unpubl.; 8Curtis et al. 1994; 9Menkis et al. 2005; 10Managbanag et al. unpubl.; 11Crous et al. 2004; 12Wirsel et al. 2002. All others are newly reported here.
Cultivation and microscopy
For growth rate determination and phenetic description of colonies, strains were point inoculated on potato-dextrose agar (PDA, Difco), OA and Blakeslee malt extract agar (MEA, Samson et al. 2002) and incubated at 25 °C for 14 d in darkness. Surface colours were rated using the colour charts of Kornerup & Wanscher (1967). For studies of microscopic morphology, strains were grown on synthetic nutrient agar (SNA, Gams et al. 2007) in slide cultures. SNA blocks of approximately 1 × 1 cm were cut out aseptically, placed upon sterile microscope slides, and inoculated at the upper four edges by means of a conidial suspension (Pitt 1979). Inoculated agar blocks were covered with sterile cover slips and incubated in moist chambers for 7 d at 25 °C in darkness. The structure and branching pattern of conidiophores were observed at magnifications × 100, × 200 and × 400 in intact slide cultures under the microscope without removing the cover slips from the agar blocks. For higher magnifications (× 400, × 1 000) cover slips were carefully removed and mounted in lactic acid with aniline blue.
Morphological parameters
Morphological terms follow David (1997), Kirk et al. (2001) and Schubert et al. (2007 - this volume). Conidiophores in Cladosporium are usually ascending and sometimes poorly differentiated. Though the initiation point of conidiophore stipes could sometimes be determined only approximately, their lengths were in some cases useful for distinguishing morphologically similar species when observed in slide cultures. The branching patterns can be rotationally symmetric or unilateral. Characters of conidial scars were studied by light and scanning electron microscopy (SEM). Conidial chains show different branching patterns, determined by the numbers of conidia in unbranched parts, the nature of ramoconidia as well as their distribution in conidial chains. Measurements are given as (i) n1-n2 or (ii) (n1-)n3-n4(-n2), with n1 = minimum value observed; n2 = maximum value observed; n3/n4 = first/third quartile. For conidia and ramoconidia also average values and standard deviations are listed. The values provided are based on at least 25 measurements for the conidiophores of each strain, and at least 50 measurements for conidia.
Ecophysiology
To determine the degree of halotolerance, strains were point-inoculated on MEA without and with additional NaCl at concentrations of 5, 10, 17 and 20 % NaCl (w/v) and incubated at 25 °C for 14 d. To determine cardinal temperature requirements for growth, plates were incubated at 4, 10, 25, 30 and 37 °C, and colony diameters measured after 14 d of incubation.
DNA extraction, sequencing and analysis
For DNA isolation strains were grown on MEA for 7 d. DNA was extracted according to Gerrits van den Ende & de Hoog (1999) by mechanical lysis of approx. 1 cm2 of mycelium. A fragment of the rDNA including the Internal Transcribed Spacer region 1, 5.8S rDNA and the ITS 2 (ITS) was amplified using the primers V9G (de Hoog & Gerrits van den Ende 1998) and LS266 (Masclaux et al. 1995). Sequence reactions were done using primers ITS1 and ITS4 (White et al. 1990). For amplification and sequencing of the partial actin gene, primers ACT-512F and ACT-783R were applied according to Carbone & Kohn (1999). For amplification and sequencing of the β-tubulin gene primers T1 and T22 were used according to O'Donnell & Cigelnik (1997). A BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, U.S.A.) was used in sequence reactions. Sequences were obtained with an ABI Prism 3700 DNA Analyzer (Applied Biosystems). They were assembled and edited using SeqMan v. 3.61 (DNAStar, Inc., Madison, U.S.A.). Sequences downloaded from GenBank are indicated in the trees by their GenBank accession numbers; newly generated sequences are indicated by strain numbers (see also Table 1). Sequences were automatically aligned using ClustalX v. 1.81 (Jeanmougin et al. 1998). The alignments were adjusted manually using MEGA3 (Kumar et al. 2004). Phylogenetic relationships of the taxa were estimated from aligned sequences by the maximum parsimony criterion as implemented in PAUP v. 4.0b10 (Swofford 2003). Data sets of the SSU rDNA, ITS rDNA and the β-tubulin and actin genes are analysed separately. Species of Cladosporium s. str. were compared with various taxa of the Mycosphaerellaceae using SSU rDNA sequences and Fusicladium effusum G. Winter (Venturiaceae) as outgroup. The other data sets focus on Cladosporium s. str., using Cladosporium salinae Zalar, de Hoog & Gunde-Cimerman as an outgroup, because this species was most deviant within Cladosporium in the SSU rDNA analysis (see below). Heuristic searches were performed on all characters, which were unordered and equally weighted. Gaps were treated as missing characters. Starting tree(s) were obtained via stepwise, random, 100 times repeated sequence addition. Other parameters included a “MaxTrees” setting to 9 000, the tree-bisection-reconnection as branch-swapping algorithm, and the “MulTrees” option set to active. Branch robustness was tested in the parsimony analysis by 10 000 search replications, each on bootstrapped data sets using a fast step-wise addition bootstrap analysis. Bootstrap values larger than 60 are noted near their respective branches. Newly generated sequences were deposited in GenBank (www.ncbi.nlm.nih.gov); their accession numbers are listed in Table 1. Alignments and trees were deposited in TreeBASE (www.treebase.org).
RESULTS
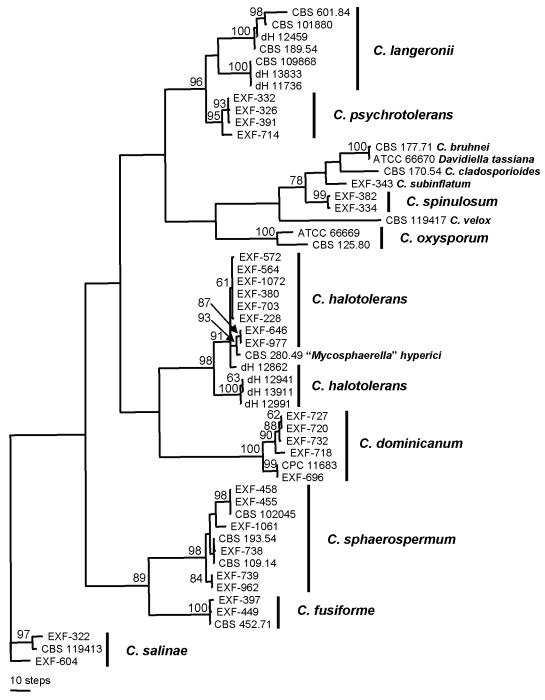
Descriptive statistical parameters of phylogenetic analyses and calculated tree scores for each analysed sequence locus are summarised in Table 2. Mainly reference material such as ex-type or ex-neotype strains was analysed on the level of SSU rDNA sequences. Downloaded and newly generated SSU rDNA sequences of members of Cladosporium s. str. were compared with related taxa of the Mycosphaerellaceae, Dothioraceae and Dothideaceae. The somewhat more distantly related Fusicladium effusum (Venturiaceae) (Braun et al. 2003: Fig. 2) was selected as outgroup. Anungitopsis amoena R.F. Castañeda & Dugan (now placed in Fusicladium Bonord., see Crous et al. 2007b), also a member of the Venturiaceae, was included in the analyses. All taxa included in the SSU rDNA analysis belong to the Dothideomycetes (Schoch et al. 2006), within which the ingroup is represented by the orders Capnodiales (Davidiellaceae, Mycosphaerellaceae, Teratosphaeriaceae) and Dothideales (Dothioraceae, Dothideaceae) (see also Schoch et al. 2006). The genus Cladosporium, of which some species are linked to Davidiella Crous & U. Braun teleomorphs (Braun et al. 2003), forms a statistically strongly supported monophyletic group (Davidiellaceae). It also accommodates species newly described in this paper, namely, C. halotolerans Zalar, de Hoog & Gunde-Cimerman, C. fusiforme Zalar, de Hoog & Gunde-Cimerman, C. dominicanum Zalar, de Hoog & Gunde-Cimerman, C. salinae, C. psychrotolerans Zalar, de Hoog & Gunde-Cimerman, C. velox Zalar, de Hoog & Gunde-Cimerman and C. spinulosum Zalar, de Hoog & Gunde-Cimerman (Fig. 1). A sister group relationship of Cladosporium s. str. with a clade of taxa characterised, among others, by Mycosphaerella Johanson teleomorphs, containing various anamorphic genera such as Septoria Sacc., Ramularia Unger, Cercospora Fresen., Pseudocercospora Speg., “Trimmatostroma” Corda (now Catenulostroma Crous & U. Braun) (see Crous et al. 2004, 2007a - this volume) and the somewhat cladosporium-like genus Devriesia Seifert & N.L. Nick. (Seifert et al. 2004), was statistically only moderately supported (Fig. 1), whereas in an analogous analysis by Braun et al. (2003: Fig. 2) it was highly supported. These data also support the conclusion by Braun et al. (2003) and Crous et al. (2006) that Cladosporium is not a member of the distantly related Herpotrichiellaceae (Chaetothyriomycetes), which is also rich in cladosporium-like taxa (Crous et al. 2006). None of the fungi isolated from hypersaline environments belonged to the Herpotrichiellaceae. The SSU rDNA sequences do not resolve a phylogenetic structure within Cladosporium s. str. Only a moderately supported clade comprising C. halotolerans, C. dominicanum, C. velox, C. sphaerospermum and C. fusiforme is somewhat distinguished from a statistically unsupported clade with C. herbarum (Pers. : Fr.) Link, C. cladosporioides (Fresen.) G.A. de Vries, C. oxysporum Berk. & Broome, C. spinulosum, and C. psychrotolerans, etc. Because C. salinae appeared most distinct within the genus Cladosporium in analyses of the SSU rDNA (Fig. 1), it was used as outgroup in analyses of the ITS rDNA and the β-tubulin and actin genes.
Table 2.
Statistical parameters describing phylogenetic analyses performed on sequence alignments of four different loci.
| Parameter | SSU rDNA | ITS rDNA1 | β-tubulin2 | Actin3 |
|---|---|---|---|---|
| Number of alignment positions | 1031 | 498 | 654 | 210 |
| Number of parsimony informative characters (PIC) | 50 | 68 | 220 | 103 |
| Length of tree / number of steps | 103 | 102 | 714 | 338 |
| Consistency Index (CI) | 0.631 | 0.804 | 0.538 | 0.586 |
| Retention Index (RI) | 0.895 | 0.975 | 0.883 | 0.885 |
| Rescaled Consistency Index (RC) | 0.565 | 0.784 | 0.475 | 0.518 |
| Homoplasy index (HI) | 0.369 | 0.196 | 0.462 | 0.414 |
| Number of equally parsimonious trees retained | 30 | 600 | 90 | 32 |
Including the internal transcribed spacer region 1 and 2 and the 5.8S rDNA.
Including partial sequences of 4 exons and complete sequences of 3 introns.
Including partial sequences of 3 exons and 2 introns.
Fig. 2.
One of 600 equally most parsimonious and equally looking phylogenetic trees based on a heuristic tree search using aligned sequences of the internal transcribed spacer regions 1 and 2 and the 5.8S rDNA. The tree was randomly selected. Support based on 10 000 replicates of a fast step-wise addition bootstrap analysis is indicated near the branches. Trees were rooted with the strains of Cladosporium salinae. Most monophyletic species clades received high, but some deeper branches moderate, bootstrap support (CI = 0.804, RI = 0.975, PIC = 68).
Fig. 1.
One of 30 equally most parsimonious and equally looking phylogenetic trees based on a heuristic tree search using aligned small subunit ribosomal DNA sequences. The tree was randomly selected. Support based on 10 000 replicates of a fast step-wise addition bootstrap analysis is indicated near the branches. Species of Cladosporium s. str., including the seven newly described species, form a strongly supported monophyletic group among other taxa of the Mycosphaerellaceae (Dothideomycetes) (CI = 0.631, RI = 0.895, PIC = 50).
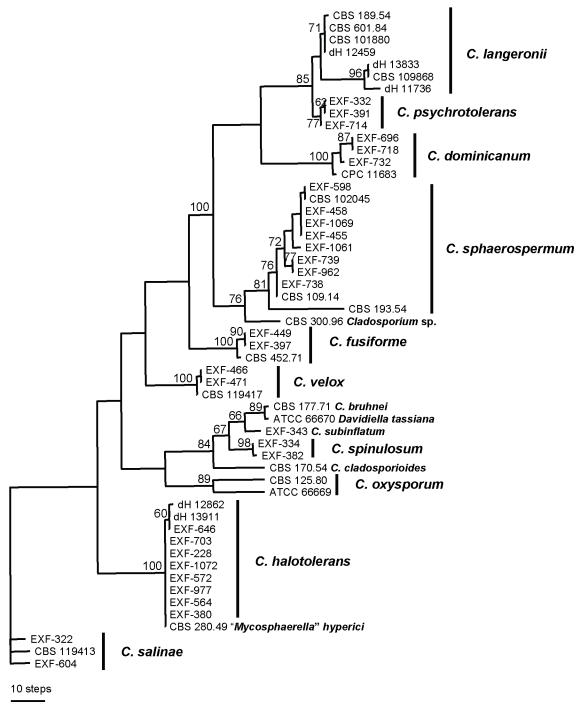
Analyses of the more variable ITS rDNA and partial β-tubulin and actin gene introns and exons supported the species clades of C. halotolerans, C. dominicanum, C. sphaerospermum, C. fusiforme and C. velox (Figs 2, 3 and 4), of which C. velox was distinguished in the β-tubulin tree by a particular long terminal branch of the only sequenced strain (Fig. 3). Cladosporium salinae also clustered as a well-supported species clade in preliminary analyses using various Mycosphaerella species as outgroup (not shown). All strains of C. langeronii (Fonseca, Leão & Nogueira) Vuill. are particularly well distinguishable from other Cladosporium species by strikingly slow-growing colonies at all tested temperatures and relatively large, oblong conidia. However, phylogenetic analyses of the β-tubulin and actin gene indicate that C. langeronii presents two cryptic species (Figs 3-4). The species clade of C. psychrotolerans is moderately supported in analyses of the actin gene but highly by means of the β-tubulin gene. It is evident from all three analyses (Figs 2, 3 and 4) that C. langeronii and C. psychrotolerans are closely related species. The species node of Cladosporium spinulosum, which is morphologically clearly distinguished from all other species by its conspicuous ornamentation consisting of digitate projections (Fig. 5), is supported by β-tubulin (Fig. 3) and actin (Fig. 4) sequence data but not by those of the ITS rDNA (Fig. 2). Analyses of all loci, however, indicate that it is a member of the C. herbarum complex.
Fig. 3.
One of 90 equally most parsimonious and equally looking phylogenetic trees based on a heuristic tree search using aligned exons and introns of a part of the β-tubulin gene. The tree was randomly selected. Support based on 10 000 replicates of a fast step-wise addition bootstrap analysis is indicated near the branches. Trees were rooted with the strains of Cladosporium salinae. Most monophyletic species clades received high, but deeper branches weak or no, bootstrap support (CI = 0.538, RI = 0.883, PIC = 220).
Fig. 4.
One of 32 equally most parsimonious and equally looking phylogenetic trees based on a heuristic tree search using aligned exons and introns of the partial actin gene. The tree was randomly selected. Support based on 10 000 replicates of a fast step-wise addition bootstrap analysis is indicated near the branches. Trees were rooted with the strains of Cladosporium salinae. Most monophyletic species clades received high, but deeper branches weak or no, bootstrap support (CI = 0.586, RI = 0.885, PIC = 103).
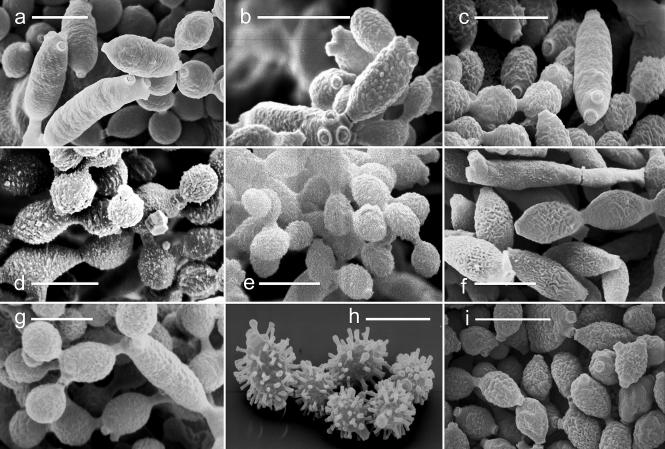
Fig. 5.
Conidial scars and surface ornamentation of ramoconidia and conidia (SEM). A. C. dominicanum (strain EXF-732). B. C. fusiforme (strain EXF-449). C. C. halotolerans (strain EXF-572). D. C. langeronii (strain CBS 189.54). E. C. psychrotolerans (strain EXF-391). F. C. salinae (strain EXF-335 = CBS 119413). G. C. sphaerospermum (strain CBS 193.54). H. C. spinulosum (strain EXF-334). I. C. velox (strain CBS 119417). Scale bars = 5 μm. (Photos: K. Drašlar).
The analyses of sequences of the ITS and the β-tubulin and actin gene introns and exons (Figs 2, 3 and 4) do not allow the full elucidation of phylogenetic relationships among these Cladosporium species. Statistical support of the interior tree branches resulting from analyses of the β-tubulin and actin genes is low (bootstrap values mostly < 50 %). While the sister group relationship of C. sphaerospermum and C. fusiforme is highly supported in the analysis based on the β-tubulin gene, analysis of the ITS rDNA indicate that these two species are unrelated, and that C. sphaerospermum is closely related to C. dominicanum. It is clear from the data that the species morphologically resembling C. sphaerospermum are not phylogenetically closely related and that the data we present here do not allow their classification in natural subgroups of the genus Cladosporium. Only C. spinulosum was placed in all analyses among species of the C. herbarum complex and all analyses supported close relatedness of C. langeronii and C. psychrotolerans.
The majority of species described here have slightly ornamented conidia ranging from minutely verruculose (C. fusiforme, C. langeronii, C. psychrotolerans, C. sphaerospermum, C. velox) to verrucose (C. halotolerans) (Fig. 5). The verrucose conidia of C. halotolerans can be recognised also under the light microscope and used as a distinguishing character. Almost smooth to minutely verruculose conidia are encountered in C. dominicanum and C. salinae (Fig. 5). Cladosporium spinulosum, a member of the C. herbarum species complex, has conidia with a digitate ornamentation that can appear spinulose under the light microscope; however, when using the SEM it became clear that its projections have parallel sides and a blunt end (Fig. 5).
DISCUSSION
The genus Cladosporium was established by Link (1816) who originally included four species, of which C. herbarum is the type species of the genus (Clements & Shear 1931). In 1950, von Arx reported a teleomorph connection for this species with Mycosphaerella tassiana (De Not.) Johanson. Based on SSU rDNA data the majority of Mycosphaerella species, including the type species of the genus, M. punctiformis (Pers.) Starbäck, clustered within the Mycosphaerellaceae, a family separated from M. tassiana (Braun et al. 2003). Therefore, Mycosphaerella tassiana was reclassified as Davidiella tassiana (De Not.) Crous & U. Braun, the type of the new genus Davidiella. All anamorphs with a cladosporium- and heterosporium-like appearance and with a supposed Dothideomycetes relationship were maintained under the anamorph name Cladosporium, morphologically characterised by scars with a protuberant hilum consisting of a central dome surrounded by a raised rim (David 1997).
The concept of distinguishing ramoconidia from secondary ramoconidia has been adopted from Schubert et al. (2007). In the species described here, ramoconidia have been observed often in C. sphaerospermum, sometimes in C. psychrotolerans, C. langeronii and C. spinulosum, and only sporadically in all other species. Therefore, ramoconidia can be seen as important for distinguishing species although sometimes, they can be observed only with difficulty. When using ramoconidia as a diagnostic criterion, colonies only from SNA and not older than 7 d should be taken into account.
Cladosporium sphaerospermum was described by Penzig (1882) from decaying Citrus leaves and branches in Italy. He described C. sphaerospermum as a species with (i) branched, septate and dark conidiophores having a length of 150-300 μm and a width of the main conidiophore stipe of 3.5-4 μm, (ii) spherical to ellipsoid, acrogenously formed conidia of 3.4-4 μm diam, and (iii) ramoconidia of 6-14 × 3.5-4 μm. Penzig's original material is not known to be preserved. Later, a culture derived from CBS 193.54, originating from a human nail, was accepted as typical of C. sphaerospermum. However, de Vries (1952), incorrectly cited it as “lectotype”, and thus the same specimen is designated as neotype in this study (see below), with the derived culture (CBS 193.54) used as ex-neotype strain. Numerous strains with identical or very similar ITS rDNA sequences as CBS 193.54 were isolated from hypersaline water or organic substrata including plants or walls of bathrooms. It is not clear yet whether surfaces in bathrooms and of plants, colonised by C. sphaerospermum, can have a similar low water activity as salterns. In our experiments, the strains of this species, however, grew under in vitro conditions at a water activity of up to 0.860, while Hocking et al. (1994) and Aihara et al. (2002) reported that it can grow even at 0.815. Therefore, we consider C. sphaerospermum as halo- or osmotolerant. Hardly any reports are available unambiguously proving that C. sphaerospermum is a human pathogen. It is therefore possible that CBS 193.54 was not involved in any disease process but rather occurred as a contaminant on dry nail material. Cladosporium sphaerospermum is a phylogenetically well-delineated species (Figs 2, 3 and 4).
Strains of C. halotolerans were isolated sporadically from substrata such as peanut cell suspension, tissue culture, bathroom walls and as culture contaminants. This surprising heterogeneity of substrata suggests that C. halotolerans is distributed by air and that it can colonise whatever substrata available, although it may have its natural niche elsewhere. We have recurrently isolated it from hypersaline water of salterns and other saline environments and it was also detected with molecular methods (but not isolated) from skin of a salt water dolphin. There are only few reports of this species from plants (Table 1). It is therefore possible that C. halotolerans is a species closely linked to salty or hypersaline environments although additional sampling is necessary to prove that. Cladosporium halotolerans is morphologically recognisable by relatively oblong to spherical, coarsely rough-walled conidia. The ITS rDNA sequence of a fungus in the skin of a bottlenose dolphin, suffering from lobomycosis, is identical to the sequences of C. halotolerans. This sequence was deposited as Lacazia loboi Taborda, V.A. Taborda & McGinnis (GenBank AF035674) by Haubold et al. (1998), who apparently concluded wrongly that a fungus with a cladosporium-like ITS rDNA sequence similar to that of C. halotolerans can be the agent of lobomycosis. Later, Herr et al. (2001) showed that Lacazia loboi phylogenetically belongs to the Onygenales on the basis of amplified SSU rDNA and chitin synthase-2 gene sequences generated from tissue lesions. By this, they confirmed an earlier supposition by Lacaz (1996) who reclassified the organism as Paracoccidioides loboi O.M. Fonseca & Silva Lacaz (Onygenales). It is therefore possible that C. halotolerans was not the main etiologic agent for the lobomycosis and it was colonising the affected dolphin skin secondarily while inhabiting other seawater habitats.
Cladosporium langeronii and C. psychrotolerans are closely related but C. langeronii is particularly well distinguishable from all other Cladosporium species by its slow growing colonies (1-7 mm diam/14 d) and relatively large conidia (4-5.5 × 3-4 μm). Cladosporium psychrotolerans has smaller conidia (3-4 × 2.5-3 μm) but a similar length : width ratio and faster expanding colonies (8-18 mm diam/14 d). Cladosporium langeronii is most likely a complex of at least two species. Strains isolated from the Arctic and the Antarctic may need to be distinguished from C. langeronii s. str. on species level. This inference is particularly supported by analyses of the β-tubulin and actin genes (Figs 3-4). Cladosporium langeronii s. str., represented by an authentic strain of Hormodendrum langeronii Fonseca, Leão & Nogueira, CBS 189.54 (Trejos 1954), has been isolated from a variety of substrata but is tolerating only up to 10 % NaCl. It was originally described by da Fonseca et al. (1927a, b) and subsequently reclassified as Cladosporium langeronii by Vuillemin (1931). The authentic strain derived from an ulcerating nodular lesion on the arm of a human patient. Because other strains of this species are ubiquitous saprobes originating from various substrata, we suspect that C. langeronii is not an important human pathogen. Cladosporium psychrotolerans has been isolated from hypersaline environments only, and tolerates up to 20 % NaCl in culture media.
In general, the human- or animal-pathogenic role of the C. sphaerospermum-like species described here seems to be limited. It is possible that pathogenic species of Cladophialophora Sacc. have been misidentified as C. sphaerospermum or as other species of Cladosporium (de Hoog et al. 2000). Alternatively, true Cladosporium species isolated as clinical strains could have been secondary colonisers since they are able to dwell on surfaces poor in nutrients, possibly in an inconspicuous dormant phase and may then be practically invisible. More likely, they could be air-borne contaminations of lesions, affected nails etc. (Summerbell et al. 2005) or are perhaps disseminated by insufficiently sterilised medical devices, as melanised fungi can be quite resistant to disinfectants (Phillips et al. 1992). They can easily be isolated and rapidly become preponderant at isolation and thus difficult to exclude as etiologic agents of a disease. For example, in 2002, a case report on an intrabronchial lesion by C. sphaerospermum in a healthy, non-asthmatic woman was described (Yano et al. 2002), but we judge the identification of the causal agent to remain uncertain, as it was based on morphology alone and no culture is available. The present authors have the opinion that all clinical cases ascribed to Cladosporium species need careful re-examination.
General characteristics and description of Cladosporium sphaerospermum-like species
The present paper focuses on Cladosporium strains isolated from hypersaline environments. Comparison of data from deliberate sampling and analysis of reference strains from culture collections inevitably leads to statistical bias, and therefore a balanced interpretation of ecological preferences of the species presented is impossible. Nevertheless, some species appeared to be consistent in their choice of habitat, and for this reason we summarise isolation data for all species described. Strains belonging to a single molecular clade proved to have similar cultural characteristics and microscopic morphology. Although within most of the species there was some molecular variation noted (particularly when intron-rich genes were analysed), some consistent phenetic trends could be observed.
Conidiophores of all C. sphaerospermum-like species lack nodose inflations (McKemy & Morgan-Jones 1991). They are usually ascending and can sometimes be poorly differentiated from their supporting hyphae. Though the initiation point of conidiophore stipes could sometimes be determined only approximately, their lengths were in some cases useful for distinguishing morphologically similar species when observed in slide cultures. Generally, the branched part of a conidiophore forms a complex tree-like structure. The number and orientation of early formed secondary ramoconidia, however, determines whether it is rotationally symmetric or unilateral.
The variability in ITS rDNA sequences observed in all C. sphaerospermum-like species (about 10 %) spans the variation observed in all members of the genus Cladosporium sequenced to date. Thus, the C. sphaerospermum-like species described here may not present a single monophyletic group but may belong to various species complexes within Cladosporium. Verifying existing literature with sequence data of these species (Wirsel et al. 2002, Park et al. 2004), we noticed that names of the common saprobes seem to be distributed nearly at random over phylogenetic trees. For most commonly used names, no type material is available for sequencing. Also verification of published reports is difficult without available voucher strains.
Cladosporium cladosporioides was incorrectly lectotypified based on CBS 170.54 (de Vries 1952), which Bisby considered a standard culture of C. herbarum. The C. cladosporioides species complex requires revision, and will form the basis of a future study. Cladosporium herbarum is maintained as a dried specimen in the Leiden herbarium; Prasil & de Hoog (1988) selected CBS 177.71 as a representative living strain. Strains, earlier accepted as living representatives of C. herbarum, CBS 177.71 and CBS 812.71 (Prasil & de Hoog 1988, Wirsel et al. 2002) and ATCC 66670 (Braun et al. 2003, as Davidiella tassiana) have been re-identified as C. bruhnei Linder by Schubert et al. (2007 - this volume). Ho et al. (1999) used strain ATCC 38027 as a representative of C. tenuissimum Cooke and this strain has identical ITS sequences as the non-deposited C. tenuissimum material used by Moricca et al. (1999). We tentatively accept this concept although we could not include ATCC 38027 in our analyses. The ITS sequence of strain CBS 125.80, identified by Wirsel et al. (2002) as C. oxysporum, is identical to the sequence of ATCC 38027. Strain ATCC 76499, published by Ho et al. (1999) as C. oxysporum, appears to be identical to a number of currently unidentified Cladosporium strains from Slovenian salterns that compose a cluster separate from all remaining species. Strains of this cluster, represented in Fig. 2 by strain ATCC 76499, morphologically resemble C. oxysporum.
Strain CBS 300.96 has not been identified to species level in the present study. It clusters outside the species clade of C. sphaerospermum, with the latter being its nearest relative. CBS 300.96 differs from C. sphaerospermum by having smaller structures: conidiophore stipes [(5-)20-80(-150) × (2-)2.5-3(-4) μm], 0-1 septate ramoconidia [(13-)19-27(-32) × 2-2.5 μm], conidia [(2.5-)3-3.5(-4) × (2-)2-2.5(-3) μm] and secondary ramoconidia [(5-)9-18(-30) × (2-)2.5-2.5(-3) μm]. However, based on a single isolate, we currently refrain from describing it as a new species.
Key to species treated in this study
Macro-morphological characters used in the key are from colonies grown on PDA and MEA 14 d at 25 °C, if not stated otherwise; microscopical characters are from SNA slide cultures grown for 7 d at 25 °C.
1. Conidial ornamentation conspicuously echinulate/digitate because of up to 1.3 μm long projections that have more or less parallel sides............................................................................................................................................................................................ C. spinulosum
1. Conidial ornamentation verruculose to verrucose or smooth, not conspicuously echinulate or digitate................................................... 2
2. Conidiophores micronematous, poorly differentiated, once or several times geniculate-sinuous, short, up to 60 μm long; terminal conidia obovoid....................................................................................................................................................................................... C. salinae
2. Conidiophores micro- or macronematous, not geniculate or only slightly so, usually up to 100 μm or 220 μm long or even longer; terminal conidia globose, subglobose to ovoid or fusiform..................................................................................................................................... 3
3. Secondary ramoconidia 0-3(-4)-septate; septa of conidiophores and conidia darkened and thickened................................................. 4
3. Secondary ramoconidia 0-1(-2)-septate; septa neither darkened nor thickened..................................................................................... 5
4. Conidiophores (5-)10-50(-300) × (2-)2.5-3(-5.5) μm; terminal conidia (2-)3-4(-6) × (2-)2.5-3(-5) μm; secondary ramoconidia (5-)7-12(-37.5) × (2-)2.5-3(-6.5) μm; ramoconidia sporadically formed.................................................................................. C. halotolerans
4. Conidiophores mostly longer and somewhat wider, (10-)45-130(-300) × (2.5-)3-4(-6) μm; terminal conidia mostly wider, (2.5-)3-4(-7) × (2-)3-3.5(-4.5) μm; secondary ramoconidia (4-)8.5-16(-37.5) × (2-)3-3.5(-5) μm; ramoconidia often formed, up to 40 μm long, with up to 5 septa............................................................................................................................................................. C. sphaerospermum
5. Terminal conidia usually fusiform........................................................................................................................................... C. fusiforme
5. Terminal conidia globose, subglobose or ovoid......................................................................................................................................... 6
6. Conidia and secondary ramoconidia irregularly verruculose to sometimes loosely verrucose; radial growth on PDA at 25 °C after 14 d typically less than 5 mm........................................................................................................................................................ C. langeronii
6. Conidia and secondary ramoconidia smooth to minutely verruculose; radial growth on PDA at 25 °C after 14 d typically more than 10 mm................................................................................................................................................................................................................... 7
7. Conidiophores (3-)3.5-4(-7.5) μm wide, thick-walled; conidiogenous loci and conidial hila 0.5-2 μm diam; ramoconidia sometimes formed with a broadly truncate, up to 2 μm wide non-cladosporioid base; no growth observed after 14 d at 30 °C on MEA.................................................................................................................................................................................... C. psychrotolerans
7. Conidiophores mostly narrower, 2-4 μm wide, only with slightly thickened walls; conidiogenous loci and conidial hila narrower, 0.5-1.5 μm diam; ramoconidia rarely formed; colony showing at least weak growth after 14 d at 30 °C on MEA................................................ 8
8. Secondary ramoconidia (4-)6.5-13(-24.5) μm long; no visible colony growth after 14 d at 10 °C on MEA.................... C. dominicanum
8. Secondary ramoconidia mostly longer, (3.5-)5.5-19(-42) μm; radial growth of colonies after 14 d at 10 °C on MEA more than 5 mm....................................................................................................................................................................................................... C. velox
Description of Cladosporium species
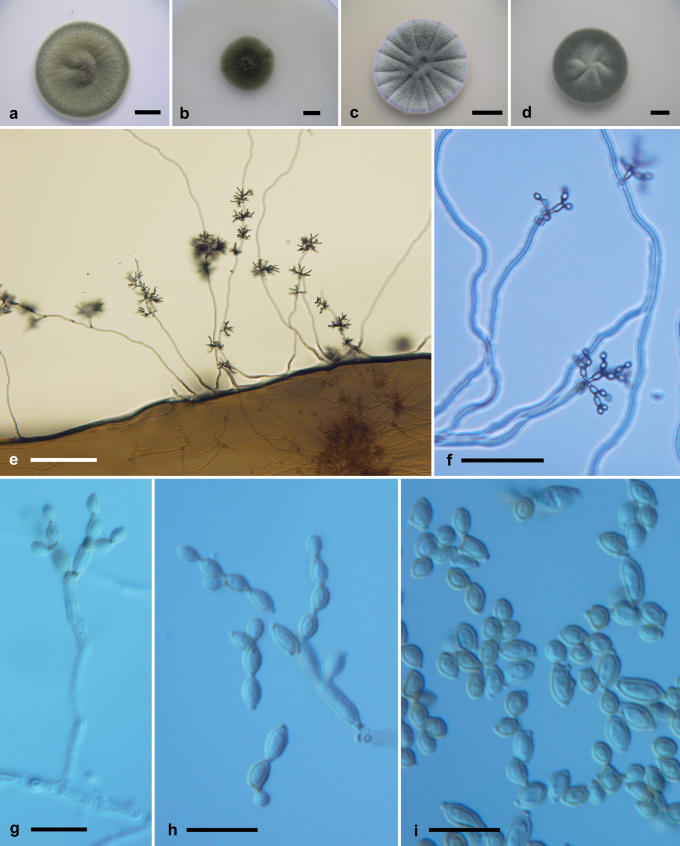
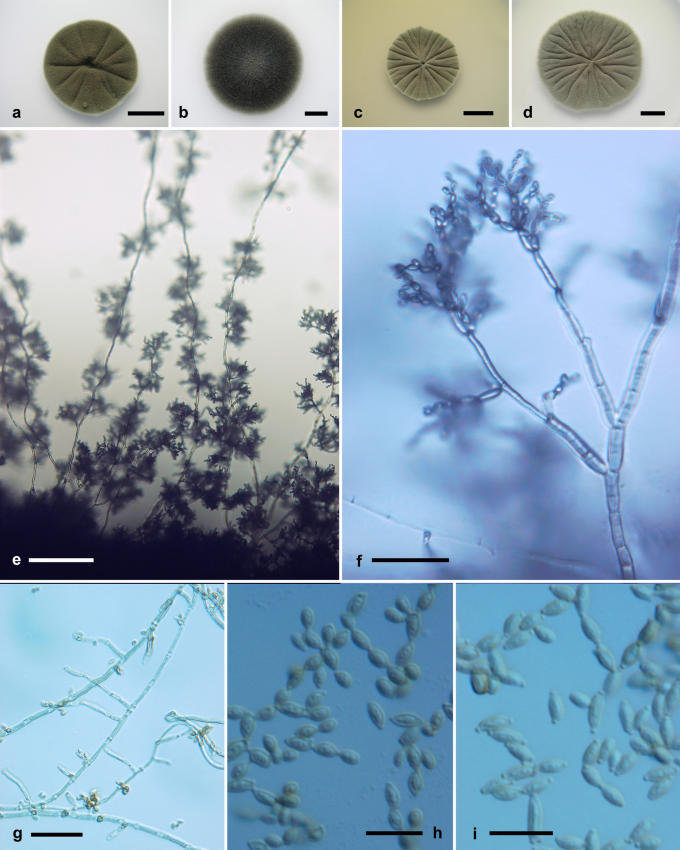
Cladosporium dominicanum Zalar, de Hoog & Gunde-Cimerman, sp. nov. MycoBank MB510995. Fig. 6.
Fig. 6.
Cladosporium dominicanum. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-F. Habit of conidiophores. G. Conidiophore. H-I. Secondary ramoconidia and conidia. E-I. All from 7-d-old SNA slide cultures. A, D, F-H, from EXF-2519; B, C, E from EXF-727; I, EXF-732 (ex-type strain). Scale bars A-D = 10 mm, E = 100 μm, F = 30 μm, G-I = 10 μm.
Etymology: Refers to the the Dominican Republic, where most strains were encountered.
Conidiophora lateralia vel terminalia ex hyphis rectis oriunda; stipes longitudine variabili, (5-)10-100(-200) × (1.5-)2-2.5(-3.5) μm, olivaceo-brunneus, levis vel leniter verruculosus, tenuitunicatus, plerumque unicellularis, simplex vel ramosus. Conidiorum catenae undique divergentes, ad 8 conidia in parte continua continentes. Cellulae conidiogenae indistinctae. Conidia levia vel leniter verruculosa, dilute brunnea, unicellularia, plerumque breviter ovoidea, utrinque angustata, (2.5-)3-3.5(-5.5) × (2-)2-2.5(-2.5) μm, long.: lat. 1.4-1.6; ramoconidia secundaria cylindrica vel quasi globosa, 0-1-septata, (4-)6.5-13(-24.5) × (2-)2.5-3(-4.5) μm, ad 4 cicatrices terminales ferentia; cicatrices inspissatae, protuberantes, 0.5-1.2 μm diam. Hyphae vagina polysaccharidica carentes.
Mycelium without extracellular polysaccharide-like material. Conidiophores arising laterally and terminally on erect hyphae, micronematous and semimacronematous, stipes of variable length, (5-)10-100(-200) × (1.5-)2-2.5(-3.5) μm, olivaceous-brown, smooth to minutely verruculose, thin-walled, almost non-septate, unbranched or branched. Conidial chains branching in all directions, up to eight conidia in the unbranched parts. Conidiogenous cells undifferentiated. Ramoconidia rarely formed. Conidia smooth to minutely verruculose, subhyaline to light brown, non-septate, usually short-ovoid, narrower at both ends, length : width ratio = 1.4-1.6; (2.5-)3-3.5(-5.5) × (2-)2-2.5(-2.5) μm [av. (± SD) 3.4 (± 0.6) × 2.2 (± 0.2)]; secondary ramoconidia cylindrical to almost spherical, 0-1-septate, (4-)6.5-13(-24.5) × (2-)2.5-3(-4.5) μm [av. (± SD) 10.3 (± 5.2) × 2.7 (± 0.6)], with up to four distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.5-1.2 μm diam.
Cultural characteristics: Colonies on PDA reaching 18-36 mm diam, olive-yellow (2D6), hairy granular, flat or slightly furrowed, with flat margin. Droplets of light reseda-green (2E6) exudate sometimes present. Reverse dark green to black. Colonies on OA reaching 19-34 mm diam, olive (2F5), loosely powdery with raised central part due to fasciculate bundles of conidiophores. Reverse dark green. Colonies on MEA reaching 30-32 mm diam, reseda green (2E6), velvety, furrowed, with undulate margin. Reverse dark green-brown. Colonies on MEA + 5 % NaCl reaching 37-41 mm diam, reseda-green (2E6), radially furrowed, velvety, sporulating in the central part or all over the colony, margin white and regular. Reverse brownish green.
Maximum tolerated salt concentration: 75 % of tested strains develop colonies at 20 % NaCl after 7 d, while after 14 d all strains grow and sporulate.
Cardinal temperatures: No growth at 4 and 10 °C, optimum 25 °C (30-32 mm diam), maximum 30 °C (2-15 mm diam), no growth at 37 °C.
Specimen examined: Dominican Republic, from hypersaline water of salt lake Enriquillo, coll. Nina Gunde-Cimerman, Jan. 2001, isol. P. Zalar 25 Feb. 2001, CBS H-19733, holotype, culture ex-type EXF-732 = CBS 119415.
Habitats and distribution: Fruit surfaces; hypersaline waters in (sub)tropical climates.
Differential parameters: No growth at 10 °C, ovoid conidia, large amounts of sterile mycelium.
Strains examined: CPC 11683, EXF-696, EXF-718, EXF-720, EXF-727, EXF-732 (= CBS 119415; ex-type strain).
Note: Cultures of C. dominicanum sporulate less abundantly than C. sphaerospermum and C. halotolerans and tend to lose their ability to sporulate with subculturing.
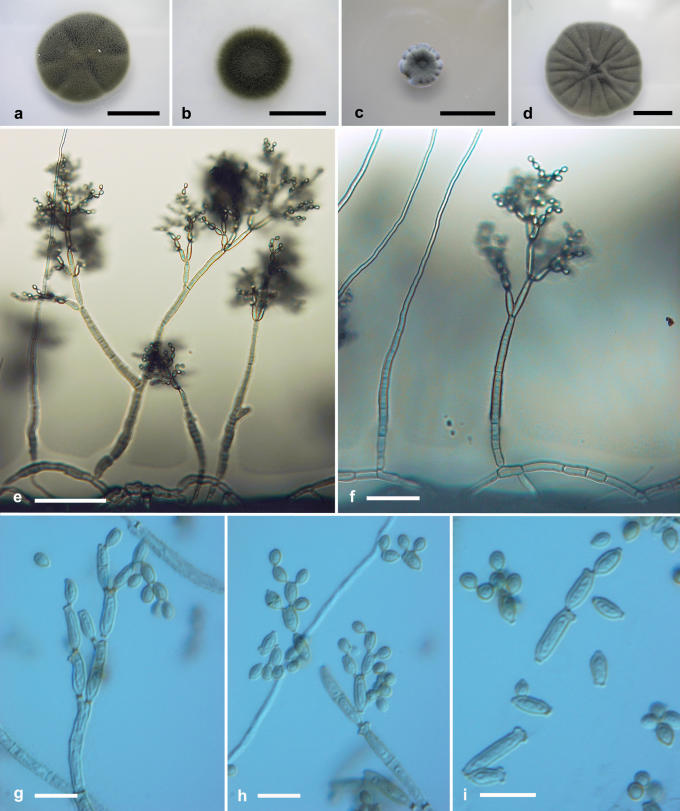
Cladosporium fusiforme Zalar, de Hoog & Gunde-Cimerman, sp. nov. MycoBank MB510997. Fig. 7.
Fig. 7.
Cladosporium fusiforme. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-G. Habit of conidiophores. H-I. Ramoconidia and conidia. E-I. All from 7-d-old SNA slide cultures. A-H, from EXF-449 (ex-type strain); I, from CBS 452.71. Scale bars A-D = 10 mm, E = 100 μm, F-G = 30 μm, H-I = 10 μm.
Etymology: Refers to its usually fusiform conidia.
Conidiophora erecta, lateralia vel terminalia ex hyphis rectis oriunda; stipes longitudine variabili, (10-)25-50(-100) × (2-)2-3.5(-4) μm, olivaceo-brunneus, levis, crassitunicatus, compluries septatus (cellulis 9-23 μm longis), plerumque simplex. Conidiorum catenae undique divergentes, in parte continua ad 5 conidia continentes. Cellulae conidiogenae indistinctae. Conidia leniter verruculosus, dilute brunnea, unicellularia, plerumque fusiformia, utrinque angustata, (2.5-)3.5-5(-6.5) × (2-)2-2.5(-3) μm, long.: lat. 1.8-2.0; ramoconidia secundaria cylindrica, 0(-1)-septata, (5-)6-11(-22) × (2.5-)2.5-3(-3) μm, ad 4 cicatrices terminales ferentia; cicatrices inspissatae, conspicuae, 0.7-1.0 μm diam. Hyphae vagina polysaccharidica carentes.
Mycelium without extracellular polysaccharide-like material. Conidiophores erect, arising laterally and terminally from straight hyphae, stipes of variable length, (10-)25-50(-100) × (2-)2-3.5(-4) μm, olivaceous-brown, smooth- and thick-walled, regularly-septate (cell length 9-23 μm), mostly unbranched. Conidial chains branching in all directions, up to 5 conidia in the unbranched parts. Conidiogenous cells undifferentiated. Ramoconidia rarely formed. Conidia minutely verruculose, light brown, aseptate, usually fusiform and narrower at both ends, length : width ratio = 1.8-2.0; (2.5-)3.5-5(-6.5) × (2-)2-2.5(-3) μm [av. (± SD) 4.4 (± 0.8) × 2.2 (± 0.2)]; secondary ramoconidia cylindrical, 0(-1)-septate, (5-)6-11(-22) × (2.5-)2.5-3(-3) μm [av. (± SD) 9.0 (± 4.7) × 2.6 (± 0.3)], with up to 4 distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.7-1.0 μm diam.
Cultural characteristics: Colonies on PDA reaching 20-26 mm diam, dull green (30E3), granular due to profuse sporulation, flat, with flat margin. Sterile mycelium absent. Reverse blackish green. Colonies on OA reaching 24-28 mm diam, olive (3F3), granular in concentric circles, consisting of two kinds of conidiophores (low and high), flat, with flat margin. Reverse black. Colonies on MEA reaching 23-28 mm diam, olive (3E5), deeply furrowed, velvety (sporulating all over) with undulate, white margin. Reverse brownish green. Colonies on MEA + 5 % NaCl reaching 28-43 mm diam, olive (3E6), granular due to profuse sporulation, slightly furrowed with flat, olive-grey (3F2) margin. Reverse dark green.
Maximum tolerated salt concentration: Only one of three strains tested (CBS 452.71) developed colonies at 17 % NaCl after 14 d, the other two strains grew until 10 % NaCl.
Cardinal temperatures: For one of three strains (CBS 452.71) the minimum temperature of growth was 4 °C (6 mm diam), for the other two 10 °C (8-9 mm diam); optimum 25 °C (23-28 mm diam), maximum 30 °C (only strain CBS 452.71 grew 5 mm diam), no growth at 37 °C.
Specimen examined: Slovenia, from hypersaline water of Sečovlje salterns, coll. and isol. L. Butinar, Dec. 1999, CBS H-19732, holotype, culture ex-type EXF-449 = CBS 119414.
Habitats and distribution: Osmotic environments worldwide.
Differential parameters: Oblong conidia, relatively low degree of halotolerance.
Strains examined: CBS 452.71, EXF-397, EXF-449 (= CBS 119414; ex-type strain).
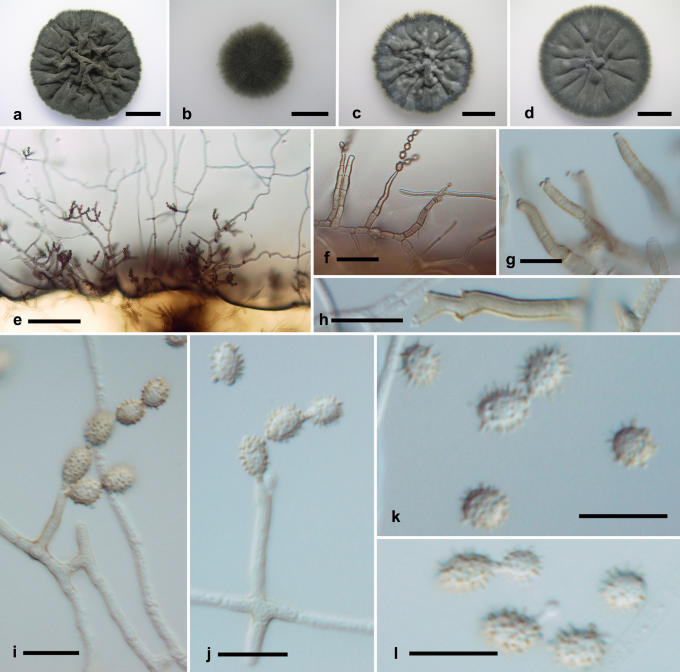
Cladosporium halotolerans Zalar, de Hoog & Gunde-Cimerman sp. nov. MycoBank MB492439. Fig. 8.
Fig. 8.
Cladosporium halotolerans. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-H. Habit of conidiophores. I. Conidiophore. J. Succession of secondary ramoconidia. K. Conidia. E-K. All from 7-d-old SNA slide cultures. A-B, from EXF-572 (ex-type strain); C-D, from EXF-977; E, G, from EXF-972; F, from EXF-564; H, I, K, from EXF-1072; J, from dH 12862. Scale bars A-D = 10 mm, E = 100 μm, F-G = 50 μm, H = 30 μm, I-K = 10 μm.
Etymology: Refers to its halotolerant habit.
Conidiophora erecta, lateralia vel terminalia ex hyphis rectis oriunda; stipes longitudine variabili, (5-)10-50(-300) × (2-)2.5-3(-5.5) μm, pallide olivaceo-brunneus, levis vel leniter verruculosus, tenuitunicatus, 0-3-septatus, interdum pluriseptatus, simplex, denticulatus. Conidiorum catenae undique divergentes, terminales ad 9 conidia continentes. Cellulae conidiogenae indistinctae. Conidia verrucosa, brunnea vel fusca, unicellularia, plerumque subglobosa vel globosa, raro breviter ovoidea, utrinque angustata, (2-)3-4(-6) × (2-)2.5-3(-5) μm, long.: lat. 1.2-1.5; ramoconidia secundaria cylindrica vel quasi globosa, 0(-1)-septata, (5-)7-12(-37.5) × (2-)2.5-3(-6.5) μm, ad 4 cicatrices terminales ferentia; cicatrices inspissatae, conspicuae, protuberantes, 0.7-1.0(-1.5) μm diam. Hyphae vagina polysaccharidica carentes. Mycelium partly submerged, partly superficial; hyphae without extracellular polysaccharide-like material. Conidiophores erect, arising laterally and terminally from straight hyphae, stipes of variable length, (5-)10-50(-300) × (2-)2.5-3(-5.5) μm, pale olivaceous-brown, smooth to minutely verruculose, thin-walled, 0-3-septate, unbranched, with pronounced denticles. Conidial chains branching in all directions, terminal chains with up to 9 conidia. Conidiogenous cells undifferentiated. Ramoconidia rarely formed. Conidia verrucose, brown to dark brown, non-septate, usually subglobose to globose, less often short-ovoid, narrower at both ends, length : width ratio = 1.2-1.5; (2-)3-4(-6) × (2-)2.5-3(-5) μm [av. (± SD) 3.5 (± 0.7) × 2.7 (± 0.5)]; secondary ramoconidia cylindrical to almost spherical, 0-1-septate, (5-)7-12(-37.5) × (2-)2.5-3(-6.5) μm [av. (± SD) 10.3 (± 4.8) × 2.9 (± 0.6)], with up to 4 distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.7-1.0(-1.5) μm diam.
Cultural characteristics: Colonies on PDA reaching 27-43 mm diam, olive (2F5), slightly furrowed, often covered with grey secondary mycelium, except at the marginal area where only sporulating structures can be observed. Margin white and regular, with submerged hyphae. Reverse pale green to black. Colonies on OA reaching 29-40 mm diam, olive (2F6), flat, uniform, granular due to profuse sporulation and fasciculate bundles of conidiophores, without sterile mycelium. Reverse dark green to black. Colonies on MEA reaching 18-44 mm diam, highly variable in colour, but mainly olive (2E5), and from flat with regular margin to deeply furrowed with undulate margin. Colony centre wrinkled with crater-shaped appearance. Reverse pale to dark green. Colonies on MEA + 5 % NaCl reaching 24-48 mm diam, olive (3E8), furrowed, velvety, with more pale, undulate margins. Reverse dark green to black.
Maximum tolerated salt concentration: Only 15 % of tested strains develop colonies at 20 % NaCl after 7 d, whereas after 14 d all cultures grow and sporulate.
Cardinal temperatures: No growth at 4 °C, optimum 25 °C (18-44 mm diam), maximum 30 °C (6-23 mm diam). No growth at 37 °C.
Specimen examined: Namibia, from hypersaline water of salterns, coll. Nina Gunde-Cimerman, 1 Sep. 2000, isol. P. Zalar, 1 Oct. 2000, CBS H-19734, holotype, culture ex-type EXF-572 = CBS 119416.
Habitats and distribution: Hypersaline water in subtropical climates; indoor environments; Arctic ice; contaminant in lesions of humans and animals; plant phyllosphere; rock.
Literature: Haubold et al. (1998), Meklin et al. (2004).
Differential parameters: Verrucose conidia, short unbranched and non-septate conidiophores which arise laterally alongside erect hyphae.
Strains examined: CBS 191.54, CBS 573.78, CBS 626.82, dH 12862, dH 12991, dH 13911, EXF-228, EXF-380, EXF-565, EXF-567, EXF-571, EXF-572 (= CBS 119416; ex-type strain), EXF-646, EXF-698, EXF-703, EXF-944, EXF-972, EXF-977, EXF-1072, EXF-2372.
Notes: Cladosporium halotolerans strongly resembles C. sphaerospermum. Several strains of this species such as dH 12862, dH 12941, CBS 191.54 and UAMH 7686 have been isolated sporadically from various indoor habitats in Europe, Brazil and the U.S.A. and repeatedly from bathrooms in Slovenia (Table 1). Probably sometimes as uncertain culture contaminations, it has been isolated from plants (GenBank accession no. L25433), inner organs of a diseased frog (AY361982) and human brain (Kantarcioglu et al. 2002). The presence of C. halotolerans species in gypsum sediments entrapped in Arctic ice, the fact that it was repeatedly isolated from hypersaline water and possibly its presence in dolphin skin (see Discussion) suggest that it has a clear preference for (hyper)osmotic habitats. This is supported by its ability to grow at 20 % NaCl.
The teleomorph of C. halotolerans is predicted to be a Davidiella species. Strain CBS 280.49 was isolated by J.A. von Arx from teleomorphic material of a fungus labelled as Mycosphaerella hyperici (Auersw.) Starbäck on Hypericum perforatum in Switzerland. According to Aptroot (2006) this species may belong in Davidiella and produces a Septoria anamorph. In the original herbarium specimen, CBS H-4867, a Mycosphaerella teleomorph was present, but no sign of a Cladosporium anamorph. We assume that CBS 280.49 was a culture contaminant.
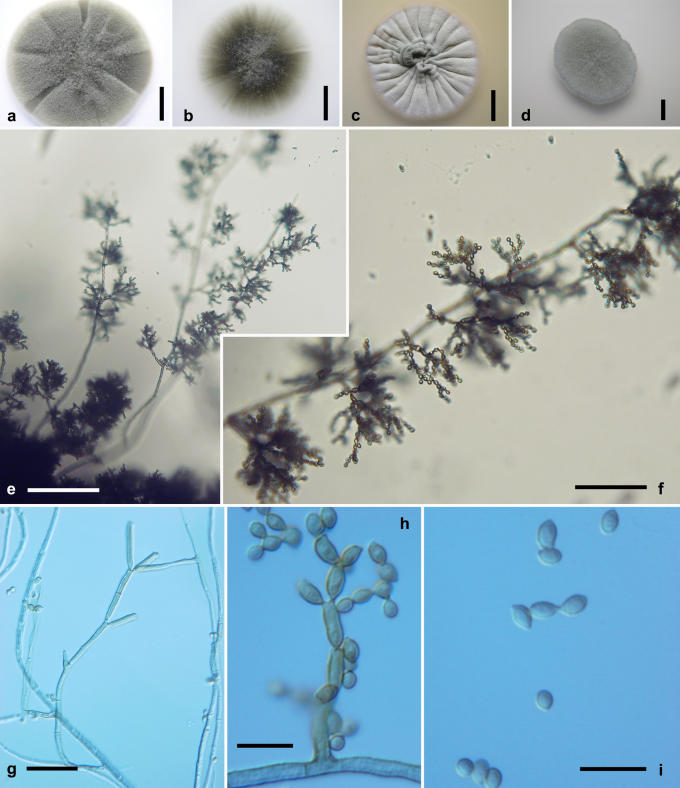
Cladosporium langeronii (Fonseca, Leão & Nogueira) Vuill., Champ. Paras.: 78. 1931. Fig. 9.
Fig. 9.
Cladosporium langeronii. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-F. Habit of conidiophores. G-I. Ramoconidia and conidia. E-I. All from 7-d-old SNA slide cultures. A-D, from CBS 189.54 (ex-type strain); E, from CBS 109868; F-I, from EXF-999. Scale bars A, C-D = 10 mm, B = 5 mm, E = 100 μm, F = 30 μm, G-I = 10 μm.
Basionym: Hormodendrum langeronii Fonseca, Leão & Nogueira, Sci. Med. 5: 563. 1927.
≡ Cladosporium langeronii (Fonseca, Leão & Nogueira) Cif., Manuale di Micologia Medica, ed. 2: 488 (1960), comb. superfl.
Mycelium partly submerged, partly superficial; hyphae sometimes enveloped in polysaccharide-like material. Conidiophores erect or ascending, micronematous and macronematous, stipes of variable length, (20-)50-130(-200) × (3-)3.5-4.5(-6.5) μm, dark brown, rough- and thick-walled, regularly septate (cell length 9-22 μm), arising laterally and terminally from submerged or aerial hyphae, branched. Conidial chains dichotomously branched, up to 6 conidia in the unbranched parts. Conidiogenous cells undifferentiated, sometimes seceding and forming ramoconidia. Ramoconidia cylindrical, 0-1 septate, (10-)11-22(-42) × (3-)3.5-4.5(-5) μm, base broadly truncate, 2-3.5 μm wide, slightly thickened and somewhat darkened. Conidia irregularly verruculose to sometimes loosely verrucose, dark brown, non-septate, usually ovoid, length : width ratio = 1.3-1.5; conidial size (3-)4-5.5(-8) × (2-)3-4(-5) μm [av. (± SD) 4.8 (± 1.0) × 3.5 (± 0.6)]; secondary ramoconidia cylindrical to almost spherical, mostly 0-1(-2)-septate, (5.5-)7.5-12.5(-35.5) × (2.5-)3-4.5(-5.5) μm [av. (± SD) 10.7 (± 4.7) × 3.6 (± 0.8)], with 2, rarely 3 distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.9-1.5(-2.3) μm diam.
Cultural characteristics: Colonies on PDA, OA and MEA with restricted growth, attaining 2.5-4.5, 1.5-7.0 and 1.0-5.5 mm diam, respectively. Colonies flat or heaped (up to 3 mm), dark green (30F4), with black reverse and slightly undulate margin with immersed mycelium. Sporulating on all media. On MEA + 5 % NaCl growth is faster, colonies attaining 8.5-12.0 mm diam, sporulating and growing deeply into the agar.
Maximum tolerated salt concentration: All strains develop colonies at 17 % NaCl after 14 d.
Cardinal temperatures: No growth at 4 °C, optimum/maximum 25 °C (1.0-5.5 mm diam), no growth at 30 °C.
Specimen examined: Brazil, from man ulcero-nodular mycosis of hand and arm, 1927, coll. and isol. da Fonseca, CBS H-19737, holotype, culture ex-type CBS 189.54.
Habitats and distribution: Polar ice and biomats; conifer wood and window frame in Europe; humans; strains originating from nasal mucus (Buzina et al. 2003) have 100 % sequence homology with the strains studied, as well as with a clone from mycorrhizal roots (Menkis et al. 2005). The species is distributed worldwide, without any apparent predilection for a particular habitat. The strains from clinical cases probably were culture contaminants.
Literature: da Fonseca et al. (1927a, b).
Differential parameters: Restricted growth; lowest salt halotolerance taxon of all C. sphaerospermum-like species.
Strains examined: CBS 189.54 (ex-type strain), CBS 601.84, CBS 101880, CBS 109868, dH 11736, dH 12459 = EXF-999, dH 13833 = EXF-1933.
Notes: De Vries (1952) synonymised the isolate identified as Hormodendrum langeronii with C. sphaerospermum. Strains of this species have often been identified as C. cladosporioides (Buzina et al. 2003, Menkis et al. 2005) although it has slightly longer conidia.
Cladosporium psychrotolerans Zalar, de Hoog & Gunde-Cimerman, sp. nov. MycoBank MB492428. Fig. 10.
Fig. 10.
Cladosporium psychrotolerans. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-F. Conidiophores. G. Apical part of a conidiophore. H-I. Secondary ramoconidia and conidia. E-I. All from 7-d-old SNA slide cultures. All but C, from EXF-391 (ex-type strain); C, from EXF-714. Scale bars A-D = 10 mm, E = 100 μm, F = 50 μm, G-I = 10 μm.
Etymology: Refers to its ability to grow at low temperatures.
Mycelium partim submersum; hyphae vagina polysaccharidica carentes. Conidiophora erecta vel adscendentia; stipes (10-)50-100(-150) × (3-)3.5-4(-7.5) μm, olivaceo-brunneus, levis, crassitunicatus, compluries regulariter septatus (cellulis 10-40 μm longis), identidem dichotome ramosus. Conidiorum catenae undique divergentes, terminales partes simplices ad 4 conidia continentes. Cellulae conidiogenae indistinctae. Ramoconidia primaria cylindrica, (18-)19-22(-43) × (2.5)3-3.5(-4.5) μm, 0(-1)-septata. Conidia leves vel leniter verruculosa, dilute brunnea, unicellularia, globosa vel ovoidea, (2.5-)3-4(-4.5) × (2-)2.5-3(-3) μm, long.: lat. 1.3-1.4; ramoconidia secundaria cylindrica, 0-1(-2)-septata, (5-)8-16(-36) × (2-)2.5-3(-5) μm, ad 4 cicatrices terminales ferentia; cicatrices inspissatae, conspicuae, 0.5-2 μm diam.
Mycelium partly superficial partly submerged; hyphae without extracellular polysaccharide-like material. Conidiophores erect or ascending, macronematous, stipes (10-)50-100(-150) × (3-)3.5-4(-7.5) μm, olivaceous-brown, smooth or almost so, thick-walled, regularly septate (cell length 10-40 μm), arising laterally from aerial hyphae, repeatedly dichotomously branched. Conidial chains branching in all directions, up to 4 conidia in the unbranched parts. Ramoconidia sometimes formed, cylindrical, (18-)19-22(-43) × (2.5)3-3.5(-4.5) μm, aseptate, rarely 1-septate, with a broadly truncate base, up to 2 μm wide, unthickened or slightly thickened, somewhat darkened-refractive. Conidia smooth to minutely verruculose, light brown, non-septate, spherical to ovoid, length : width ratio = 1.3-1.4; conidial size (2.5-)3-4(-4.5) × (2-)2.5-3(-3) μm [av. (± SD) 3.4 (± 0.5) × 2.5 (± 0.2)]; secondary ramoconidia cylindrical, 0-1(-2)-septate, (5-)8-16(-36) × (2-)2.5-3(-5) μm [av. (± SD) 12.7 (± 6.5) × 3.0 (± 0.5)], with up to 4 distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.5-2 μm diam.
Cultural characteristics: Colonies on PDA reaching 13-18 mm diam, velvety, olive (3F4) due to profuse sporulation, flat with straight margin. Reverse dark green. Colonies on OA reaching 13-15 mm diam, olive (2F8), of granular appearance due to profuse sporulation; aerial mycelium sparse. Margin regular. Reverse black. Colonies on MEA reaching 8-15 mm diam, olive (2F4), velvety, radially furrowed with undulate white margin. Colonies on MEA with 5 % NaCl growing faster than on other media, reaching 25-27 mm diam, olive (3E6) and granular due to profuse sporulation, either slightly furrowed or heavily wrinkled with regular or undulate margin. Reverse dark green.
Maximum tolerated salt concentration: 17 % NaCl after 14 d.
Cardinal temperatures: Minimum at 4 °C (5 mm diam), optimum and maximum at 25 °C (8-15 mm diam).
Specimen examined: Slovenia, from hypersaline water of Sečovlje salterns, coll. and isol. S. Sonjak, May 1999, CBS H-19730, holotype, culture ex-type EXF-391 = CBS 119412.
Habitats and distribution: Hypersaline water in the Mediterranean basin.
Differential parameters: Growth at 4 °C; maximal NaCl concentration 17 % NaCl, which differentiates it from other species with similar conidia, like C. sphaerospermum, C. halotolerans and C. dominicanum.
Strains examined: EXF-326, EXF-332, EXF-391 (= CBS 119412; ex-type strain), EXF-714.
Cladosporium salinae Zalar, de Hoog & Gunde-Cimerman, sp. nov. MycoBank MB492438. Fig. 11.
Fig. 11.
Cladosporium salinae. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-F. Habit of conidiophores. G. Conidiophore. H. Detail of apical part of conidiophore. I. Conidia. J. Secondary ramoconidia and conidia. E-J. All from 7-d-old SNA slide cultures. A-D, from EXF-604; E-J, from EXF-335 (ex-type strain). Scale bars A-C = 5 mm, D = 10 mm, E = 100 μm, F = 50 μm, G = 30 μm, H-J = 10 μm.
Etymology: Refers to salterns (= Latin salinae) as the habitat of this species.
Mycelium partim submersum; hyphae multa rostra lateralia ferentes, hyphae vagina polysaccharidica involutae. Conidiophora vix distincta, lateralia vel terminalia ex hyphis aeriis oriunda; stipes longitudine variabili, (5-)25-50(-60) × (2-)2.5-3(-4) μm, olivaceo-brunneus, levis vel leniter verruculosus, crassitunicatus, irregulariter dense septatus (cellulis 6-29 μm longis), simplex, interdum ramosus. Conidiorum catenae undique divergentes, terminales ad 6 conidia continentes. Cellulae conidiogenae nonnumquam integratae, in summo sequentiam sympodialem denticulorum formantes. Conidia levia, interdum leniter verruculosa, dilute brunnea, unicellularia, plerumque fusiformia, (4.5-)5.5-7.5(-10) × (2-)2.5-3(-3.5) μm, long.: lat. 1.9-2.4; ramoconidia secundaria cylindrica, 0-1(-2)-septata, (7.5-)9.5-13.5(-19) × (2.5-)2.5-3.5(-4.5) μm, ad 5 cicatrices terminales ferentia; cicatrices inspissatae, conspicuae, protuberantes, 0.7-1.8 μm diam.
Mycelium partly superficial partly submerged, with numerous lateral pegs, consistently enveloped in polysaccharide-like material. Conidiophores poorly differentiated, micronematous, stipes (5-)25-50(-60) × (2-)2.5-3(-4) μm, olivaceous-brown, smooth to often minutely verruculose or irregularly rough-walled, thick-walled, irregularly densely septate (length of cells 6-29 μm), arising laterally and terminally from aerial hyphae, unbranched, occasionally branched. Conidial chains branching in all directions, terminal chains with up to 6 conidia. Conidiogenous cells sometimes integrated, producing sympodial clusters of pronounced denticles at their distal ends. Conidia usually smooth, occasionally minutely verruculose, light brown, aseptate, usually oblong ellipsoidal to fusiform, length : width ratio = 1.9-2.4; (4.5-)5.5-7.5(-10) × (2-) 2.5-3(-3.5) μm [av. (± SD) 6.7 (± 1.3) × 2.9 (± 0.4)]; secondary ramoconidia cylindrical, 0-1(-2)-septate, (7.5-)9.5-13.5(-19) × (2.5-)2.5-3.5(-4.5) μm [av. (± SD) 12.1 (± 3.3) × 3.2 (± 0.6)], with up to 5 distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.7-1.8 μm diam.
Cultural characteristics: Colonies on PDA reaching 10-27 mm diam, granular, olive (2E4) due to profuse sporulation, with white undulate margin. Aerial mycelium absent. Colonies either heaped or radially furrowed, in the marginal area growing deeply into the agar. Reverse dark brown to dark green. Colonies on OA reaching 7-20 mm diam, olive (3E6), of granular appearance due to profuse sporulation, aerial mycelium present. Margin either undulate or arachnoid, deeply furrowed. Reverse pale brown to dark green. Colonies on MEA reaching 8-19 mm diam, velvety, reseda-green (2E6), heaped. Margin furrowed, growing deeply into the agar. Colonies on MEA with 5 % NaCl growing much faster than on other media, reaching 25-38 mm diam, of different colours, mostly reseda-green (2E6) and granulate due to profuse sporulation, margin olive-yellow (2D6). Reverse yellow to dark green.
Maximum tolerated salt concentration: MEA + 17 % NaCl after 14 d.
Cardinal temperatures: No growth at 4 °C, optimum and maximum temperature at 25 °C (8-19 mm diam), no growth at 30 °C.
Specimen examined: Slovenia, from hypersaline water of Sečovlje salterns, coll. and isol. S. Sonjak, Feb. 1999, CBS H-19731, holotype, culture ex-type EXF-335 = CBS 119413.
Habitats and distribution: Hypersaline water in the Mediterranean basin.
Differential parameters: Sympodial conidiogenous cells with pronounced denticles, narrow temperature amplitude. Strains examined: EXF-322, EXF-335 (= CBS 119413; ex-type strain), EXF-604.
Notes: Cladosporium salinae morphologically resembles species of the genus Fusicladium because its conidia are oblong ellipsoidal to fusiform and conidiogenous loci of ramoconidia are placed closely together. As any other Cladosporium species, its conidia show typical cladosporioid scar structures, however. Cladosporium salinae seems to have a separate position within the genus Cladosporium since it seems to be distantly related to any other described Cladosporium species or currently known species complex within Cladosporium.
Cladosporium sphaerospermum Penzig, Michelia 2(8): 473. 1882. Fig. 12.
Fig. 12.
Cladosporium sphaerospermum. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-F. Habit of conidiophores. G-I. Ramoconidia and conidia. E-I. All from 7-d-old SNA slide cultures. A, C-D, F-H, from CBS 193.54 (ex-neotype strain); B, from EXF-738; E, EXF-455; I, EXF-458. Scale bars A-D = 10 mm; E = 100 μm; F = 50 μm; G-I = 10 μm.
Mycelium partly submerged, partly superficial; hyphae thick, darkly pigmented and densely septate in submerged mycelium, not enveloped in polysaccharide-like material. Conidiophores erect or ascending, micronematous and macronematous, stipes of variable length, (10-)45-130(-300) × (2.5-)3-4(-6) μm, olivaceous-brown, smooth to minutely verruculose, thick-walled, with relatively dense septation (cells mostly 4.5-23 long), septa darkened and somewhat thickened, arising laterally and terminally from immersed or aerial hyphae, either unbranched or branched. Conidial chains branching in all directions, up to 6 conidia in the unbranched parts. Conidiogenous cells not differentiated. Ramoconidia often formed, cylindrical, (11.5-)20.5-40(-48) × (2.5-)3(-3.5) μm, with up to 5 septa, base broadly truncate, 2 μm wide, slightly thickened and somewhat darkened-refractive. Conidia verruculose, brown to dark brown, non-septate, usually subspherical to spherical, less often short-ovoid, narrower at both ends, with length : width ratio = 1.1-1.5; conidial size (2.5-)3-4(-7) × (2-)3-3.5(-4.5) μm [av. (± SD) 3.8 (± 0.8) × 3.1 (± 0.4)]; secondary ramoconidia cylindrical to almost spherical, 0-3(-4) septate, (4-)8.5-16(-37.5) × (2-)3-3.5(-5) μm [av. (± SD) 13.1 (± 6.3) × 3.2 (± 0.5)], with up to 4, rarely up to 6 distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.9-1.1(-1.4) μm diam.
Cultural characteristics: Colonies on PDA reaching 21-44 mm diam, velvety, olive (2F5) due to profuse sporulation, either with white and regular, or exceptionally undulate margin. Aerial mycelium sparse. Colonies flat or rarely radially furrowed with elevated colony centre. Exudates not prominent, some strains release green soluble pigments into the agar. Reverse blackish blue to pale green. Growth deep into the agar. Colonies on OA reaching 21-38 mm diam, olive (2F8), of granular appearance due to profuse and uniform sporulation, almost no aerial mycelium. Margin either regular or arachnoid, deeply radially furrowed. Reverse black. Colonies on MEA reaching 15-35 mm diam, velvety, linden-green (2C5), radially furrowed. Colony centre wrinkled, forming a crater-like structure; margin furrowed, lighter in colour, consisting of submerged mycelium. Reverse pale to dark brown. Colonies on MEA with 5 % NaCl growing faster than on other media, reaching 31-60 mm diam, mainly olive (2D4), either being almost flat or radially furrowed, with margin of superficial mycelium; sporulation dense. Reverse ochraceous or dark green.
Maximum tolerated salt concentration: On MEA + 20 % NaCl 89 % of all strains tested develops colonies after 7 d, 96 % after 14 d.
Cardinal temperatures: No growth at 4 °C, optimum 25 °C (15-35 mm diam), maximum 30 °C (2-15 mm diam). No growth at 37 °C.
Specimen examined: Netherlands, from nail of man, 1949, coll. and isol. R.W. Zappey, CBS H-19738, neotype designated here, incorrectly selected by de Vries (1952) as “lectotype”, culture ex-neotype CBS 193.54 = ATCC 11289 = IMI 049637.
Habitats and distribution: Hypersaline water in mediterranean and tropics; soil and plants in temperate climates; indoor wet cells; humans. The species does not seem to have any particular preference. Human isolates were probably culture contaminants.
Literature: Penzig (1882), de Vries (1952), Ellis (1971), de Hoog et al. (2000), Samson et al. (2002).
Diagnostic parameters: Thick-walled, melanised, densely septate mycelium, almost spherical, verruculose to verrucose terminal conidia, growth on 20 % NaCl after 7 d.
Strains examined: CBS 109.14, CBS 122.63, CBS 190.54, CBS 192.54, CBS 193.54 (ex-neotype strain), CBS 102045, CPC 10944, EXF-131, EXF-328, EXF-385, EXF-446, EXF-455, EXF-458, EXF-461, EXF-464, EXF-465, EXF-598, EXF-644, EXF-645, EXF-649, EXF-715, EXF-738, EXF-739, EXF-781, EXF-962, EXF-965, EXF-1061, EXF-1726, EXF-1732.
Cladosporium spinulosum Zalar, de Hoog & Gunde-Cimerman, sp. nov. MycoBank MB501099. Fig. 13.
Fig. 13.
Cladosporium spinulosum. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E. Habit of conidiophores. F-J. Conidiophores. K-L. Conidia (also visible in I-J). E-L. All from 7-d-old SNA slide cultures. A-L, from EXF-334 (ex-type strain). Scale bars A-D = 10 mm, E = 100 μm, F = 30 μm, G-L = 10 μm.
Etymology: Refers to its conspicuously digitate conidia.
Conidiophora erecta vel adscendentia; stipites longitudine variabili, (15-)25-50(-155) × (2.5-)3-4(-5) μm, olivaceo-brunneus, levis, crassitunicatus, 0-6(-9)-septatus (cellulis 6-20 μm longis), ex hyphis submersis vel aeriis lateraliter vel terminaliter oriundus, simplex vel ramosus. Conidiorum catenae undique ramosae, ad 4 conidiis in partibus linearibus continuis cohaerentibus. Cellulae conidiogenae integratae vel discretae, acervos distales denticulorum conspicuorum sympodialium proferentes. Conidia echinulata vel digitata, brunnea vel fusca, continua, vulgo subglobosa vel globosa, (4.5-)5.5-7(-8) × (3-)4-4.5(-5) μm, long.: lat. = 1.1-1.6, digiti 0.6-1.3 μm longi; ramoconidia secundaria etiam digitata, cylindrica vel subglobosa, 0(-1)-septata, (6-)6.5-8(-18) × (4-)4.5-5(-5.5) μm, 1-3 cicatrices distales ferentia. Cicatrices inspissatae, conspicuae, protuberantes, 0.8-1.2 μm diam. Hyphae nonnumquam polysaccharido circumdatae.
Hyphae sometimes enveloped in polysaccharide-like material. Conidiophores erect or ascending, stipes of variable length, (15-) 25-50(-155) × (2.5-)3-4(-5) μm, olivaceous-brown, smooth, sometimes irregularly rough-walled to verrucose near the base, thick-walled, 0-6(-9)-septate (cells mostly 6-20 μm long), arising laterally and terminally from immersed or aerial hyphae, either unbranched or branched, somewhat tapering towards the apex. Conidial chains branching in all directions, up to 4 conidia in the unbranched parts. Conidiogenous cells sometimes integrated, producing sympodial clusters of pronounced denticles at their distal ends. Ramoconidia rarely formed. Conidial wall ornamentation conspicuously digitate, with up to 1.3 μm long projections having parallel sides and blunt ends. Conidia brown to dark brown, aseptate, usually subspherical to spherical, length : width ratio = 1.1-1.6; conidial size (4.5-)5.5-7(-8) × (3-)4-4.5(-5) μm [av. (± SD) 6.2 (± 1.0) × 4.2 (± 0.5)]; secondary ramoconidia ornamented as conidia, cylindrical to almost spherical, 0(-1)-septate, (6-)6.5-8(-18) × (4-)4.5-5(-5.5) μm [av. (± SD) 8.6 (± 4.0) × 4.8 (± 0.4)], with up to 3 distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.8-1.2 μm diam.
Cultural characteristics: Colonies on PDA reaching 20-30 mm diam, velvety, dull green (29E4) to dark green (29F6) due to profuse sporulation, either with white and regular, or undulate margin. Aerial mycelium sparse. Colonies flat or radially furrowed with elevated colony centre. Growth deep into the agar. Exudates not prominent. Colonies on OA reaching 20-25 mm diam, dull green (29E4) to dark green (29F6), sometimes olive (3D4), of granular appearance due to profuse and uniform sporulation; almost without aerial mycelium. Margin arachnoid. Reverse pale brown to black. Colonies on MEA reaching 17-28 mm diam, velvety, dull green (29E4) to dark green (29F6), either flat or radially furrowed. Colony centre wrinkled, forming a crater-like structure; margin furrowed, paler in colour, consisting of submerged mycelium only. Reverse pale to dark green. Colonies on MEA with 5 % NaCl reaching 12-18 mm diam, of different colours, greenish grey (29D2), greyish green (29D5) to dark green (29F6); colony appearance variable, mostly either being almost flat with immersed colony centre or radially furrowed, with white to dark green margin consisting of superficial mycelium; sporulation dense. Reverse pale to dark green.
Maximum tolerated salt concentration: On MEA + 17 % NaCl, two of three strains tested developed colonies after 14 d.
Cardinal temperatures: Growth at 4 °C, optimum and maximum at 25 °C (17-28 mm). No growth at 30 °C.
Specimen examined: Slovenia, from hypersaline water of Sečovlje salterns, coll. and isol. S. Sonjak, Feb. 1999, CBS H-19796, holotype, culture ex-type EXF-334 = CBS 119907.
Habitats and distribution: Hypersaline water in temperate climate.
Diagnostic parameters: Conidia and ramoconidia with a digitate ornamentation.
Strains examined: EXF-334 (= CBS 119907; ex-type strain), EXF-382.
Notes: Cladosporium spinulosum is a member of the C. herbarum species complex (Figs 2, 3 and 4) although its globoid conidia are reminiscent of C. sphaerospermum. Within Cladosporium, the species is unique in having conspicuously digitate conidia and ramoconidia. The two strains are differing in the size of conidia. The average size of conidia in EXF-334 is 6.2 (± 0.9) × 4.2 (± 0.5) μm, and in EXF-382 it is 3.9 (± 0.6) × 3.3 (± 0.4) μm.
Cladosporium velox Zalar, de Hoog & Gunde-Cimerman, sp. nov. MycoBank MB492435. Fig. 14.
Fig. 14.
Cladosporium velox. Macro- and micromorphological characters. A-D. Colony surface grown on PDA (A), OA (B), MEA (C) and MEA plus 5 % NaCl (D) of strains incubated for 14 d at 25 °C in darkness. E-F. Habit of conidiophores. G. Conidiophore. H-I. Secondary ramoconidia and conidia. E-I. All from 7-d-old SNA slide cultures. A-D, G, from CBS 119417 (ex-type strain); E-F, H-I, from EXF-466. Scale bars A-D = 10 mm, E = 100 μm, F = 50 μm, G = 30 μm, H-I = 10 μm.
Etymology: Refers to the quick growth of strains of this species.
Mycelium partim submersum; hyphae vagina polysaccharidica carentes. Conidiophora erecta, lateralia vel terminalia ex hyphis aeriis oriunda; stipes (10-) 25-150(-250) × (2.5-)3-4(-4.5) μm, olivaceo-brunneus, levis, crassitunicatus, ad 7-septatus (cellulis 10-60 μm longis), identidem dichotome ramosus. Conidiorum catenae undique divergentes, terminales partes simplices ad 5 conidia continentes. Cellulae conidiogenae indistinctae. Conidia levia vel leniter verruculosa, dilute brunnea, unicellularia, ovoidea, (2-)3-4(-5.5) × (1.5-)2-2.5(-3) μm, long.: lat. 1.4-1.7; ramoconidia secundaria cylindrica, 0-1-septata, (3.5-)5.5-19(-42) × (2-)2.5-3(-4.5) μm, ad 4(-5) cicatrices terminales ferentia; cicatrices inspissatae, protuberantes, conspicuae, 0.5-1.5 μm diam.
Mycelium partly superficial partly submerged; hyphae without extracellular polysaccharide-like material. Conidiophores erect, stipes (10-)25-150(-250) × (2.5-)3-4(-4.5) μm, slightly attenuated towards the apex, olivaceous-brown, smooth- and thick-walled, arising terminally and laterally from aerial hyphae, dichotomously branched [up to 5(-7)-septate, cell length 10-60 μm]. Ramoconidia rarely formed. Conidial chains branching in all directions, terminal chains with up to 5 conidia. Conidia smooth to very finely verruculose, pale brown, non-septate, ovoid, length : width ratio = 1.4-1.7; (2-)3-4(-5.5) × (1.5-)2-2.5(-3) μm [av. (± SD) 3.6 (± 0.6) × 2.3 (± 0.2)]; secondary ramoconidia cylindrical, 0-1-septate, (3.5-)5.5-19(-42) × (2-)2.5-3(-4.5) μm [av. (± SD) 13.4 (± 10.2) × 2.8 (± 0.5)], with up to 4(-5) distal scars. Conidiogenous scars thickened and conspicuous, protuberant, 0.5-1.5 μm diam.
Cultural characteristics: Colonies on PDA reaching 35-45 mm diam, velvety, dark green due to profuse sporulation, on some parts covered with white sterile mycelium, flat with straight white margin. Reverse dark green to black. Colonies on OA reaching 30-43 mm diam, dark green, mycelium submerged, aerial mycelium sparse. Margin regular. Reverse black. Colonies on MEA reaching 30-42 mm diam, pale green, radially furrowed, with raised, crater-shaped central part, with white, undulate, submerged margin. Sporulation poor. Colonies on MEA with 5 % NaCl reaching 35-45 mm diam, pale green, velvety, flat with regular margin. Reverse pale green. Sporulation poor.
Maximum tolerated salt concentration: 20 % NaCl after 14 d.
Cardinal temperatures: Minimum at 10 °C (9 mm diam), optimum at 25 °C (30-42 mm diam) and maximum at 30 °C (5-18 mm diam).
Specimen examined: India, Charidij, isolated from Bambusa sp., W. Gams, CBS H-19735, holotype, culture ex-type CBS 119417.
Habitats and distribution: Hypersaline water in Slovenia; bamboo, India.
Strains examined: CBS 119417 (ex-type strain), EXF-466, EXF-471.
Acknowledgments
The authors are grateful to Walter Gams for his comments on the manuscript and for providing Latin diagnoses, and to Konstanze Schubert for contributions to species descriptions. We thank Kazimir Drašlar and Marko Lutar for preparing SEM illustrations, and Kasper Luijsterburg, Špela Štrekelj and Barbara Kastelic-Bokal for their technical assistance. The research was partially supported by the European Union-funded Integrated Infrastructure Initiative grant SYNTHESYS Project NL-TAF-1070.
Taxonomic novelties: Cladosporium dominicanum Zalar, de Hoog & Gunde-Cimerman, sp. nov., C. fusiforme Zalar, de Hoog & Gunde-Cimerman, sp. nov., C. halotolerans Zalar, de Hoog & Gunde-Cimerman, sp. nov., C. psychrotolerans Zalar, de Hoog & Gunde-Cimerman, sp. nov., C. salinae Zalar, de Hoog & Gunde-Cimerman, sp. nov., C. spinulosum Zalar, de Hoog & Gunde-Cimerman, sp. nov., C. velox Zalar, de Hoog & Gunde-Cimerman, sp. nov.
References
- Aihara M, Tanaka T, Ohta T, Takatori K (2002). Effect of temperature and water activity on the growth of Cladosporium sphaerospermum and Cladosporium cladosporioides. Biocontrol Science 7: 193-196. [Google Scholar]
- Aihara M, Tanaka T, Takatori K (2001). Cladosporium as the main fungal contaminant of locations in dwelling environments. Biocontrol Science 6: 49-52. [Google Scholar]
- Aptroot A (2006). Mycosphaerella and its anamorphs: 2. Conspectus of Mycosphaerella. CBS Biodiversity Series 5: 1-231. [Google Scholar]
- Arx JA von (1950). Über die Ascusform von Cladosporium herbarum (Pers.) Link. Sydowia 4: 320-324. [Google Scholar]
- Badillet G, Bièvre C de, Spizajzen S (1982). Isolement de dématiées à partir d'ongles et de squames. Bulletin de la Société Française de Mycologie 11: 69-72. [Google Scholar]
- Braun U, Crous PW, Dugan F, Groenewald JZ, Hoog GS de (2003). Phylogeny and taxonomy of cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycological Progress 2: 3-18. [Google Scholar]
- Butinar L, Sonjak S, Zalar P, Plemenitaš A, Gunde-Cimerman N (2005). Melanized halophilic fungi are eukaryotic members of microbial communities in hypersaline waters of solar salterns. Botanica Marina 48: 73-79. [Google Scholar]
- Buzina W, Braun H, Freudenschuss K, Lackner A, Stammberger H (2003). Fungal biodiversity as found in nasal mucus. Medical Mycology 41: 149-161. [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553-556. [Google Scholar]
- Clements FE, Shear CL (1931). The genera of fungi. New York: H.W. Wilson Co.
- Crous PW, Braun U, Groenewald JZ (2007a). Mycosphaerella is polyphyletic. Studies in Mycology 58: 1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Pongpanich K, Himaman W, Arzanlou M, Wingfield MJ (2004). Cryptic speciation and host specificity among Mycosphaerella spp. occurring on Australian Acacia species grown as exotics in the tropics. Studies in Mycology 50: 457-469. [Google Scholar]
- Crous PW, Schubert K, Braun U, Hoog GS de, Hocking AD, Shin H-D, Groenewald JZ (2007b). Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Studies in Mycology 58: 185-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ (2006). Eucalyptus microfungi known from culture. 1. Cladoriella and Fulvoflamma genera nova, with notes on some other poorly known taxa. Studies in Mycology 55: 53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Gore J, Oliver RP (1994). The phylogeny of the tomato leaf mould fungus Cladosporium fulvum syn. Fulvia fulva by analysis of rDNA sequences. Current Genetics 25: 318-322. [DOI] [PubMed] [Google Scholar]
- David JC (1997). A contribution to the systematics of Cladosporium. Revision of the fungi previously referred to Heterosporium. Mycological Papers 172: 1-157. [Google Scholar]
- Ellis MB (1971). Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew.
- Fonseca FOOR da, Area Leão AE de, Peñido NJJC (1927a). Mycose de type ulcéro-nodulaire, semblable à la sporotrichose et produite par Hormodendrum langeronii. Comptes-rendus des Séances de la Société de Biologie 97: 1772-1774. [Google Scholar]
- Fonseca FOOR da, Area Leão AE de, Peñido NJJC (1927b). Mycose de type ulcéro-nodulaire, semlable à la sporotrichose et produite par Hormodendrum langeronii. Sciencia Medica 5: 563-573. [Google Scholar]
- Gams W, Verkleij GJM, Crous PW (2007). CBS Course of Mycology, 5th ed. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands.
- Gerrits van den Ende AHG, Hoog GS de (1999). Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Studies in Mycology 43: 151-162. [Google Scholar]
- Gunde-Cimerman N, Zalar P, Hoog GS de, Plemenitaš A (2000). Hypersaline water in salterns - natural ecological niches for halophilic black yeasts. FEMS Microbiology Ecology 32: 235-240. [DOI] [PubMed] [Google Scholar]
- Haubold EM, Aronson JF, Cowan DF, McGinnis MR, Cooper CR (1998). Isolation of fungal rDNA from bottlenose dolphin skin infected with Loboa loboi. Medical Mycology 36: 263-267. [PubMed] [Google Scholar]
- Herr RA, Tarcha EJ, Taborda PR, Taylor JW, Ajello L, Mendoza L (2001). Phylogenetic analysis of Lacazia loboi places this previously uncharacterised pathogen within the dimorphic Onygenales. Journal of Clinical Microbiology 39: 309-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MHM, Castañeda RF, Dugan FM, Jong SC (1999). Cladosporium and Cladophialophora in culture: descriptions and an expanded key. Mycotaxon 72: 115-157. [Google Scholar]
- Hocking AD, Miscamble BF, Pitt J (1994). Water relations of Alternaria alternata, Cladosporium cladosporioides, Cladosporium sphaerospermum, Curvularia lunata and Curvularia pallescens. Mycological Research 98: 91-94. [Google Scholar]
- Hoog GS de, Gerrits van den Ende AHG (1998). Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41: 183-189. [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Guarro J, Gené J, Figueras MA (2000). Atlas of Clinical Fungi, 2nd ed. Centraalbureau voor Schimmelcultures, Utrecht/Universitat Rovira i Virgili, Reus.
- Hoog GS de, Zalar P, Urzì C, Leo F de, Yurlova NA, Sterflinger K (1999). Relationships of dothideaceous black yeasts and meristematic fungi based on 5.8S and ITS2 rDNA sequence comparison. Studies in Mycology 43: 31-37. [Google Scholar]
- Jeannmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998). Multiple sequence alignment with ClustalX. Trends in Biochemical Sciences 23: 403-405. [DOI] [PubMed] [Google Scholar]
- Kantarcioglu AS, Yucel A, Hoog GS de (2002). Case report. Isolation of Cladosporium cladosporioides from cerebrospinal fluid. Mycoses 45: 500-503. [DOI] [PubMed] [Google Scholar]
- Kirk P, Cannon P, David J, Stalpers J (2001). Ainsworth and Bisby's Dictionary of the Fungi. 9th edn. Wallingford, UK. CAB International.
- Kornerup A, Wanscher JH (1967). Methuen Handbook of Colour. 2nd edn. Methuen Co., London.
- Kumar S, Tamura K, Nei M (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5: 150-163. [DOI] [PubMed] [Google Scholar]
- Lacaz CS (1996). Paracoccidioides loboi (Fonseca Filho et Arêa Leão, 1940) Almeida et Lacaz, 1948-1949. Description of the fungus in Latin. Revista Instituto Medicina Tropical São Paulo 38: 229-231. [DOI] [PubMed] [Google Scholar]
- Link HF (1816). Observationes in ordines plantarum naturales III. Magazin der Gesellschaft Naturforschender Freunde Berlin 7: 37-38. [Google Scholar]
- Masclaux F, Guého E, Hoog GS de, Christen R (1995). Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. Journal of Medical and Veterinary Mycology 33: 327-338. [DOI] [PubMed] [Google Scholar]
- McKemy JM & Morgan-Jones G (1991). Studies in the genus Cladosporium sensu lato. V. Concerning the type species, Cladosporium herbarum. Mycotaxon 42: 307-317. [Google Scholar]
- Meklin T, Haugland RA, Reponen T, Varma M, Lummus Z, Bernstein D, Wymer LJ, Vesper SJ (2004). Quantitative PCR analysis of house dust can reveal abnormal mold conditions. Journal of Environmental Monitoring 6: 615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A, Vasiliauskas R, Taylor AFS, Stenlid J, Finlay R (2005). Fungal communities in mycorrhizal roots of conifer seedlings in forest nurseries under different cultivation systems, assessed by morphotyping, direct sequencing and mycelial isolation. Mycorrhiza 16: 33-41. [DOI] [PubMed] [Google Scholar]
- Moricca S, Ragazzi A, Mitchelson KR (1999). Molecular and conventional detection and identification of Cladosporium tenuissimum on two-needle pine rust aeciospores. Canadian Journal of Botany 77: 339-347. [Google Scholar]
- O'Donnell K, Cigelnik E (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetic Evolution 7: 103-116. [DOI] [PubMed] [Google Scholar]
- Park HG, Managbanag JR, Stamenova EK, Jong SC (2004). Comparative analysis of common indoor Cladosporium species based on molecular data and conidial characters. Mycotaxon 89: 441-451. [Google Scholar]
- Penzig AJO (1882). Fungi Agromiculi. Michelia 2: 385-503. [Google Scholar]
- Pereira PT, Carvalho MM de, Girio FM, Roseiro JC, Amaral-Collaco MT (2002). Diversity of microfungi in the phylloplane of plants growing in a Mediterranean ecosystem. Journal of Basic Microbiology 42: 396-407. [DOI] [PubMed] [Google Scholar]
- Phillips G, Hudson S, Stewart WK (1992). Microbial growth and blockage of sub-floor drains in a renal dialysis centre: a problem highlighted. Journal of Hospital Infection 21: 193-198. [DOI] [PubMed] [Google Scholar]
- Pitt JI (1979). The Genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press, London.
- Prasil K, Hoog GS de (1988). Variability in Cladosporium herbarum. Transactions of the British Mycological Society 90: 49-54. [Google Scholar]
- Samson RA, Hoekstra ES, Frisvad JC, Filtenborg O (2002). Introduction to Food- and Airborne Fungi, 6th ed. Centraalbureau voor Schimmelcultures, Utrecht.
- Schoch C, Shoemaker RA, Seifert, KA, Hambleton S, Spatafora JW, Crous PW (2006). A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1043-1054. [DOI] [PubMed] [Google Scholar]
- Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, Zalar P, Hoog GS de, Crous PW (2007). Biodiversity in the Cladosporium herbarum (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Studies in Mycology 58: 105-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Nickerson NL, Corlett M, Jackson ED, Louis-Seize G, Davies RJ (2004). Devriesia, a new hyphomycete genus to accommodate heat-resistant, cladosporium-like fungi. Canadian Journal of Botany 82: 914-926. [Google Scholar]
- Summerbell RC, Cooper E, Bunn U, Jamieson F, Gupta AK (2005). Onychomycosis: a critical study of techniques and criteria confirming the etiologic significance of nondermatophytes. Medical Mycology 43: 39-59. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
- Trejos A (1954). Cladosporium carrionii n. sp. and the problem of cladosporia isolated from chromoblastomycosis. Revista de Biología Tropical 2: 75-112. [Google Scholar]
- Vries GA de (1952). Contribution to the knowledge of the genus Cladosporium Link ex Fr. Baarn, 121 pp. Dissertation, University of Utrecht.
- Vuillemin P (1931). Les champignons parasites et les mycoses de l'homme. Encyclopédie Mycologique 2: 1-290. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds). Academic Press, San Diego, California: 315-322.
- Wirsel SGR, Runge-Froböse C, Ahren DG, Kemen E, Oliver RP, Mendgen, KW (2002). Four or more species of Cladosporium sympatrically colonize Phragmites australis. Fungal Genetics and Biology 25: 99-113. [DOI] [PubMed] [Google Scholar]
- Yano S, Koyabashi K, Kato K (2002). Intrabronchial lesion due to Cladosporium sphaerospermum in a healthy, non-asthmatic woman. Mycoses 46: 348-350. [DOI] [PubMed] [Google Scholar]