Abstract
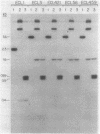
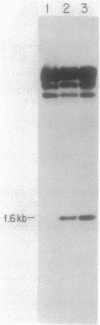
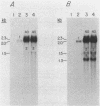
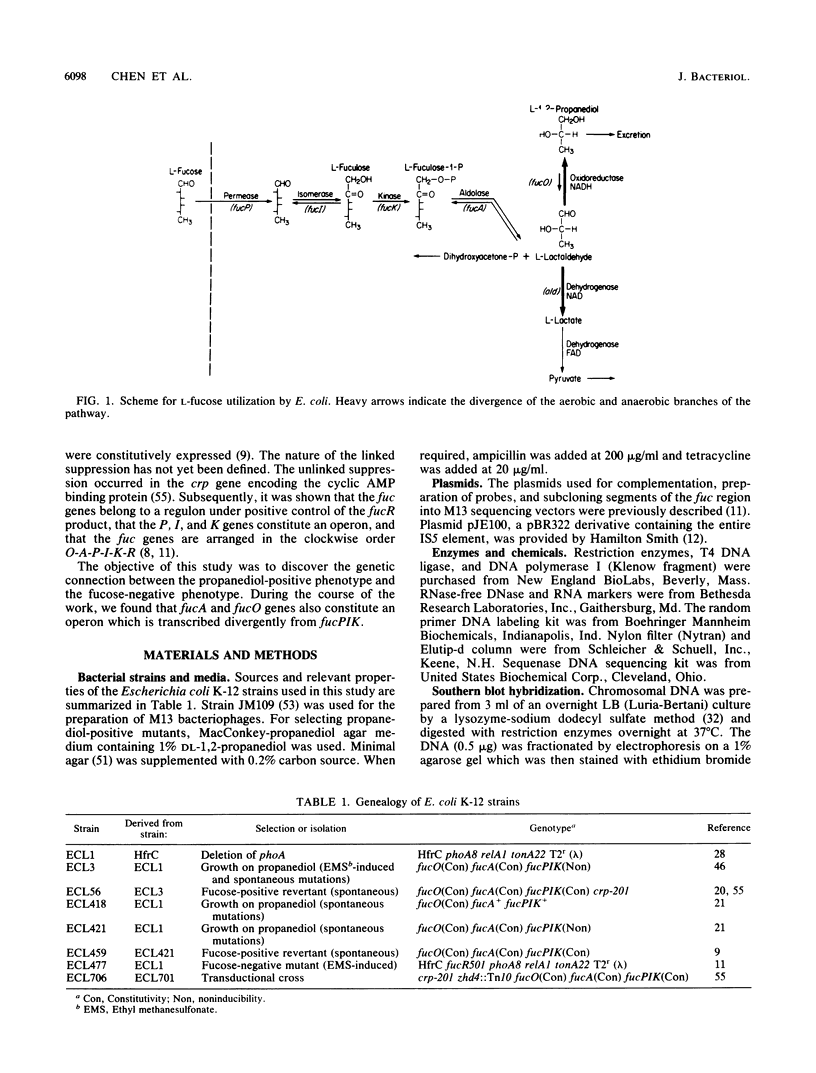
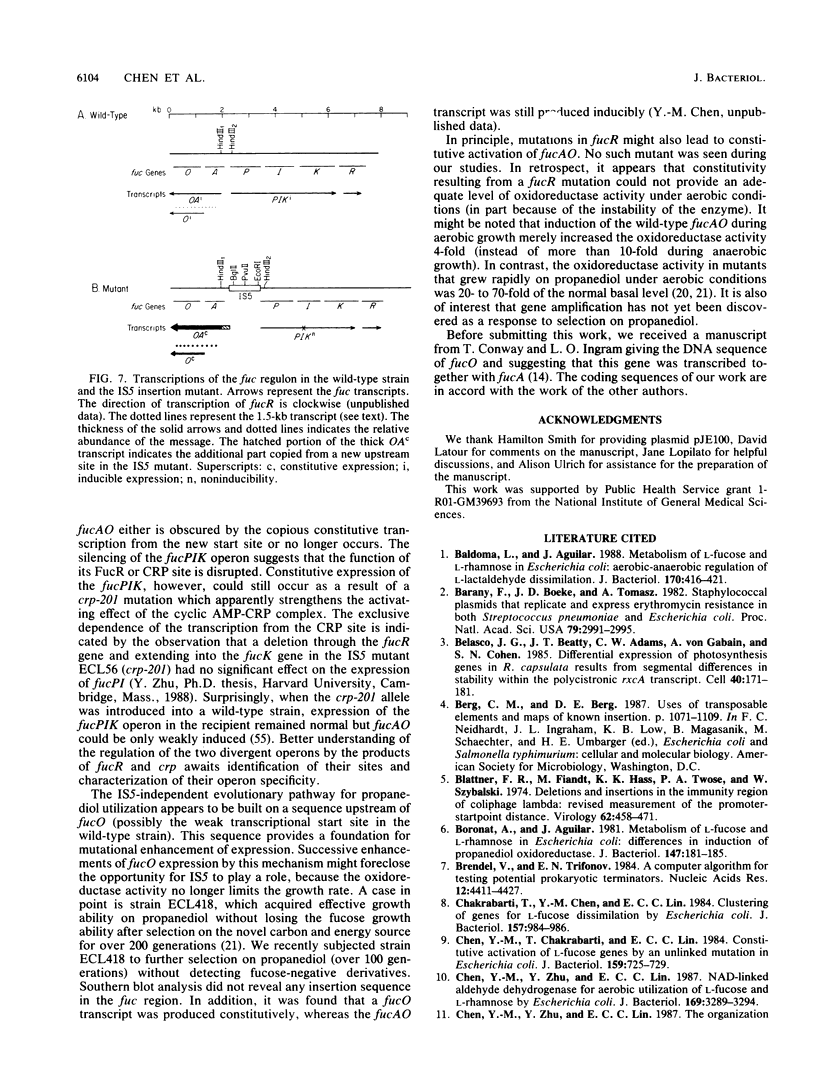
L-1,2-Propanediol is an irretrievable end product of L-fucose fermentation by Escherichia coli. Selection for increased aerobic growth rate on propanediol results in the escalation of basal synthesis of the NAD+-linked oxidoreductase encoded by fucO, a member of the fuc regulon for the utilization of L-fucose. In general, when fucO becomes constitutively expressed, two other simultaneous changes occur: the fucA gene encoding fuculose-1-phosphate aldolase becomes constitutively expressed and the fucPIK operon encoding fucose permease, fucose isomerase, and fuculose kinase becomes noninducible. In the present study, we show that fucO and fucA form an operon which is divergently transcribed from the adjacent fucPIK operon. In propanediol-positive and fucose-negative mutants the cis-controlling region shared by the operons fucAO and fucPIK is lengthened by 1.2 kilobases. DNA hybridization identified the insertion element to be IS5. This element, always oriented in the same direction with the left end (the BglII end) proximal to fucA, apparently causes constitutive expression of fucAO and noninducibility of fucPIK. The DNA of the fucAO operon and a part of the adjacent fucP was sequenced.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldomà L., Aguilar J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: aerobic-anaerobic regulation of L-lactaldehyde dissimilation. J Bacteriol. 1988 Jan;170(1):416–421. doi: 10.1128/jb.170.1.416-421.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barany F., Boeke J. D., Tomasz A. Staphylococcal plasmids that replicate and express erythromycin resistance in both Streptococcus pneumoniae and Escherichia coli. Proc Natl Acad Sci U S A. 1982 May;79(9):2991–2995. doi: 10.1073/pnas.79.9.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Boronat A., Aguilar J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: differences in induction of propanediol oxidoreductase. J Bacteriol. 1981 Jul;147(1):181–185. doi: 10.1128/jb.147.1.181-185.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti T., Chen Y. M., Lin E. C. Clustering of genes for L-fucose dissimilation by Escherichia coli. J Bacteriol. 1984 Mar;157(3):984–986. doi: 10.1128/jb.157.3.984-986.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Chakrabarti T., Lin E. C. Constitutive activation of L-fucose genes by an unlinked mutation in Escherichia coli. J Bacteriol. 1984 Aug;159(2):725–729. doi: 10.1128/jb.159.2.725-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Zhu Y., Lin E. C. NAD-linked aldehyde dehydrogenase for aerobic utilization of L-fucose and L-rhamnose by Escherichia coli. J Bacteriol. 1987 Jul;169(7):3289–3294. doi: 10.1128/jb.169.7.3289-3294.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Zhu Y., Lin E. C. The organization of the fuc regulon specifying L-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol Gen Genet. 1987 Dec;210(2):331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- Chernak J. M., Schlaffer E. J., Smith H. O. Synthesis and overproduction of the 5A protein of insertion sequence IS5. J Bacteriol. 1988 Nov;170(11):5368–5370. doi: 10.1128/jb.170.11.5368-5370.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocks G. T., Aguilar T., Lin E. C. Evolution of L-1, 2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J Bacteriol. 1974 Apr;118(1):83–88. doi: 10.1128/jb.118.1.83-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J Bacteriol. 1989 Jul;171(7):3754–3759. doi: 10.1128/jb.171.7.3754-3759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler J. A., van Bree M. P. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene. 1981 Aug;14(3):155–163. doi: 10.1016/0378-1119(81)90111-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Glansdorff N., Charlier D., Zafarullah M. Activation of gene expression by IS2 and IS3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):153–156. doi: 10.1101/sqb.1981.045.01.024. [DOI] [PubMed] [Google Scholar]
- Hacking A. J., Lin E. C. Disruption of the fucose pathway as a consequence of genetic adaptation to propanediol as a carbon source in Escherichia coli. J Bacteriol. 1976 Jun;126(3):1166–1172. doi: 10.1128/jb.126.3.1166-1172.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacking A. J., Lin E. C. Regulatory changes in the fucose system associated with the evolution of a catabolic pathway for propanediol in Escherichia coli. J Bacteriol. 1977 May;130(2):832–838. doi: 10.1128/jb.130.2.832-838.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton D. M., Musso R. E. Transcription initiation sites within an IS2 insertion in a Gal-constitutive mutant of Escherichia coli. Nucleic Acids Res. 1982 Aug 25;10(16):5015–5031. doi: 10.1093/nar/10.16.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Normark S. Insertion of IS2 creates a novel ampC promoter in Escherichia coli. Cell. 1983 Mar;32(3):809–816. doi: 10.1016/0092-8674(83)90067-3. [DOI] [PubMed] [Google Scholar]
- Jund R., Loison G. Activation of transcription of a yeast gene in E. coli by an IS5 element. Nature. 1982 Apr 15;296(5858):680–681. doi: 10.1038/296680a0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kröger M., Hobom G. Structural analysis of insertion sequence IS5. Nature. 1982 May 13;297(5862):159–162. doi: 10.1038/297159a0. [DOI] [PubMed] [Google Scholar]
- LERNER S. A., WU T. T., LIN E. C. EVOLUTION OF A CATABOLIC PATHWAY IN BACTERIA. Science. 1964 Dec 4;146(3649):1313–1315. doi: 10.1126/science.146.3649.1313. [DOI] [PubMed] [Google Scholar]
- Lin E. C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- Mekalanos J. J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983 Nov;35(1):253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- Mortlock R. P., Fossitt D. D., Wood W. A. A basis for utlization of unnatural pentoses and pentitols by Aerobacter aerogenes. Proc Natl Acad Sci U S A. 1965 Aug;54(2):572–579. doi: 10.1073/pnas.54.2.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S., Kato M., Kohara Y., Mizuno T. Insertion sequence IS5 contains a sharply curved DNA structure at its terminus. Mol Gen Genet. 1988 Nov;214(3):433–438. doi: 10.1007/BF00330477. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Rak B., Lusky M., Hable M. Expression of two proteins from overlapping and oppositely oriented genes on transposable DNA insertion element IS5. Nature. 1982 May 13;297(5862):124–128. doi: 10.1038/297124a0. [DOI] [PubMed] [Google Scholar]
- Rak B., von Reutern M. Insertion element IS5 contains a third gene. EMBO J. 1984 Apr;3(4):807–811. doi: 10.1002/j.1460-2075.1984.tb01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Felton J., Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981 Oct 22;293(5834):625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- Reynolds A. E., Mahadevan S., LeGrice S. F., Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 1986 Sep 5;191(1):85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- Saedler H., Besemer J., Kemper B., Rosenwirth B., Starlinger P. Insertion mutations in the control region of the Gal operon of E. coli. I. Biological characterization of the mutations. Mol Gen Genet. 1972;115(3):258–265. doi: 10.1007/BF00268889. [DOI] [PubMed] [Google Scholar]
- Saedler H., Reif H. J., Hu S., Davidson N. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet. 1974;132(4):265–289. doi: 10.1007/BF00268569. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988 Oct;7(10):3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner B., Kahn M. The nucleotide sequence of IS5 from Escherichia coli. Gene. 1981 Aug;14(3):165–174. doi: 10.1016/0378-1119(81)90112-8. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhara S., Wu T. T., Chused T. M., Lin E. C. Ferrous-activated nicotinamide adenine dinucleotide-linked dehydrogenase from a mutant of Escherichia coli capable of growth on 1, 2-propanediol. J Bacteriol. 1969 Apr;98(1):87–95. doi: 10.1128/jb.98.1.87-95.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlinger P. IS elements and transposons. Plasmid. 1980 May;3(3):241–259. doi: 10.1016/0147-619x(80)90039-6. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall B. G. Topological repression of gene activity by a transposable element. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6115–6119. doi: 10.1073/pnas.81.19.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvanen M. The evolutionary implications of mobile genetic elements. Annu Rev Genet. 1984;18:271–293. doi: 10.1146/annurev.ge.18.120184.001415. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Lerner S. A., Lin E. C. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J Bacteriol. 1967 Feb;93(2):642–648. doi: 10.1128/jb.93.2.642-648.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M., Hollenberg C. P. Selective cloning of Bacillus subtilis xylose isomerase and xylulokinase in Escherichia coli genes by IS5-mediated expression. EMBO J. 1984 Nov;3(11):2555–2560. doi: 10.1002/j.1460-2075.1984.tb02173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Lin E. C. A mutant crp allele that differentially activates the operons of the fuc regulon in Escherichia coli. J Bacteriol. 1988 May;170(5):2352–2358. doi: 10.1128/jb.170.5.2352-2358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Lin E. C. Loss of aldehyde dehydrogenase in an Escherichia coli mutant selected for growth on the rare sugar L-galactose. J Bacteriol. 1987 Feb;169(2):785–789. doi: 10.1128/jb.169.2.785-789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]