Abstract
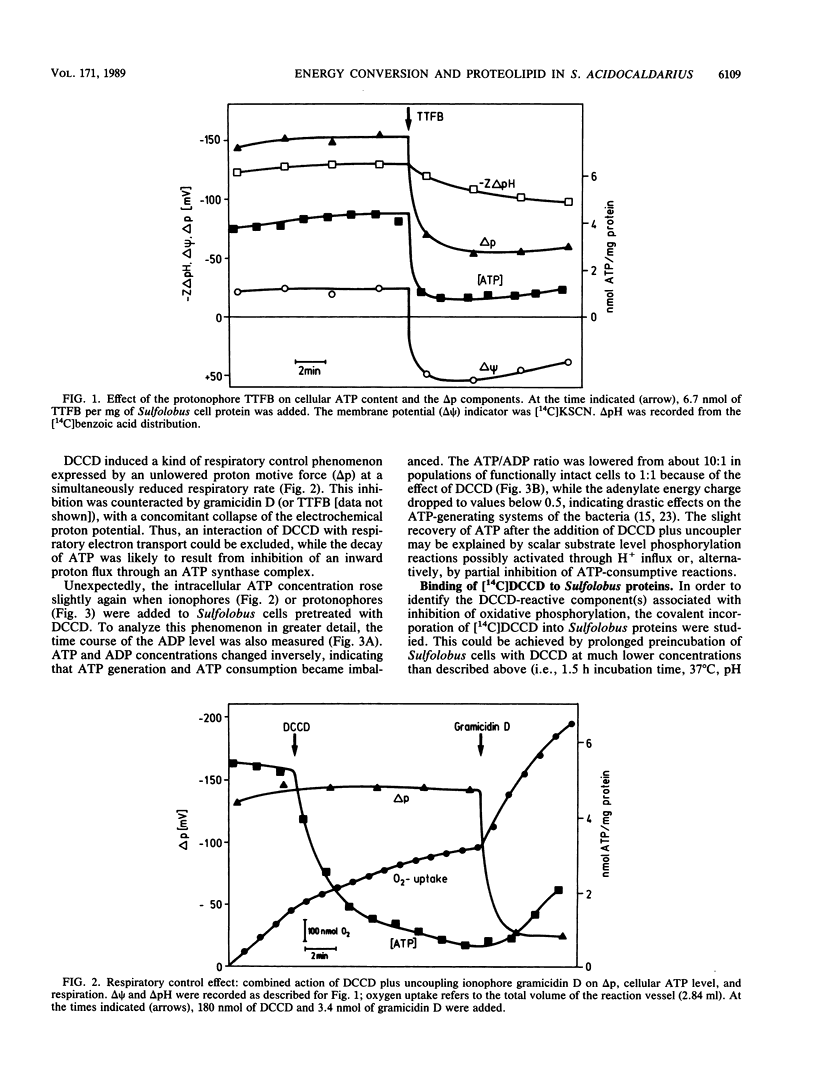
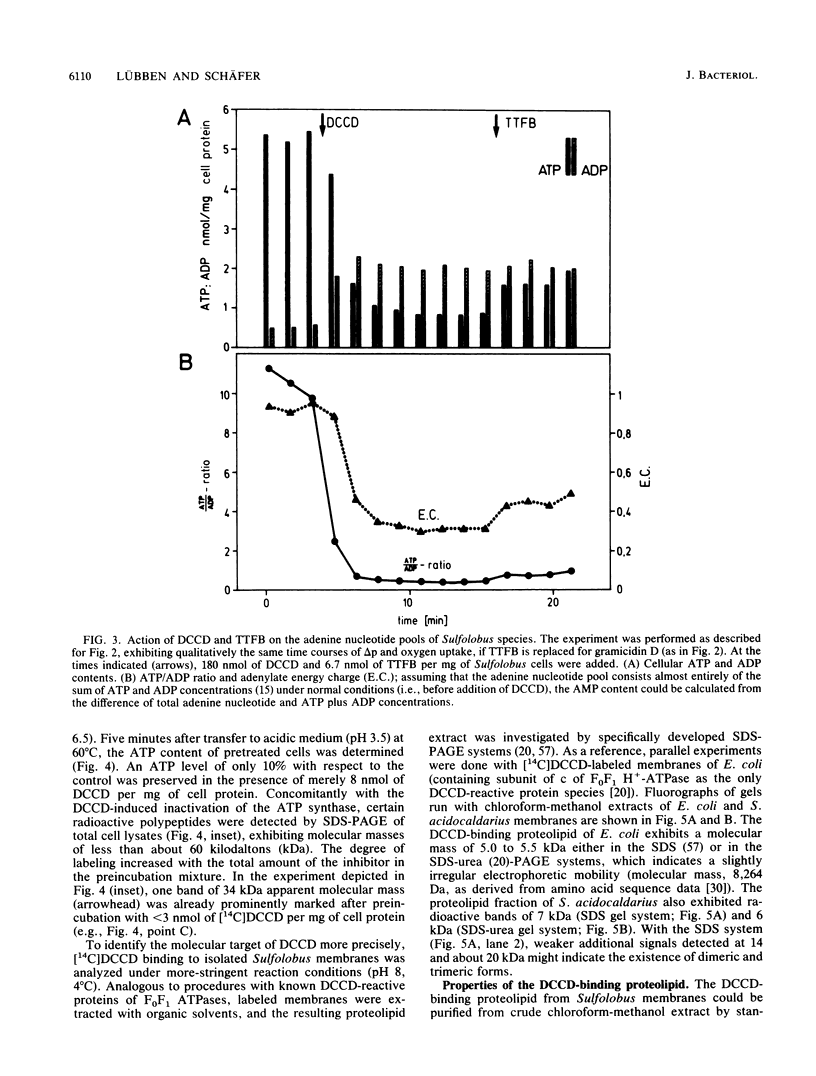
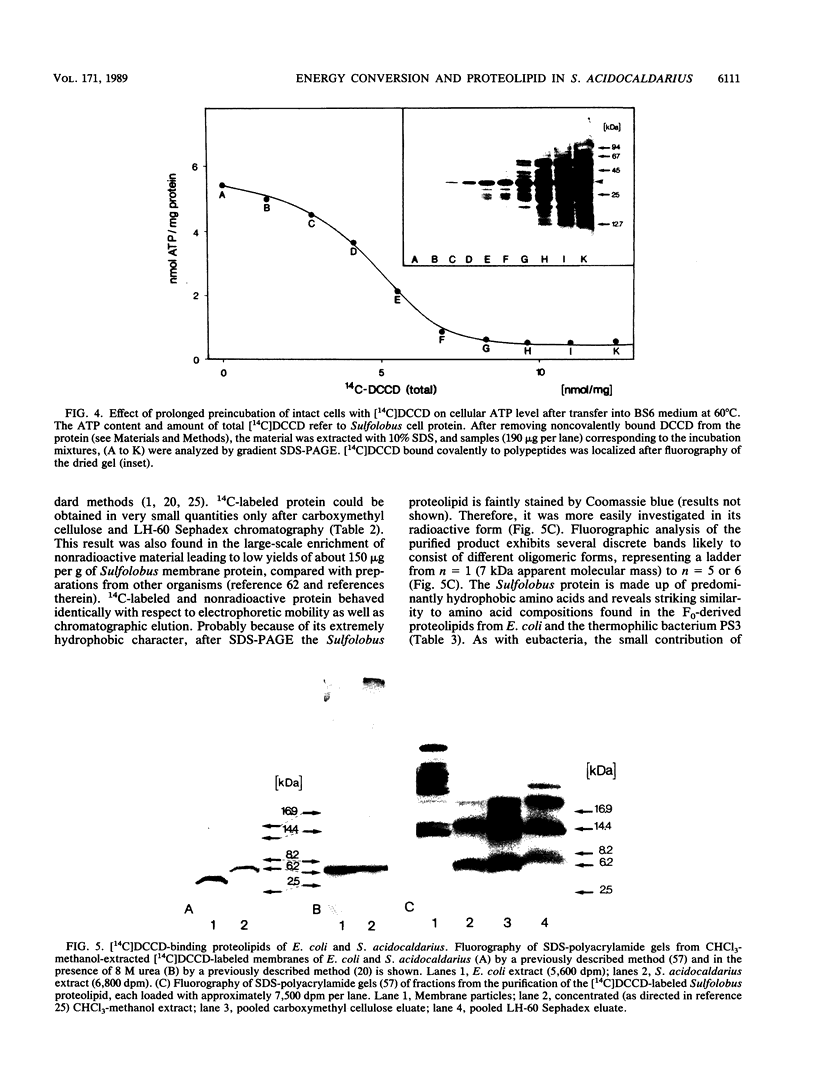
The energy-transducing mechanism of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius DSM 639 has been studied, addressing the question whether chemiosmotic proton gradients serve as an intermediate energy store driving an F0F1-analogous ATP synthase. At pH 3.5, respiring S. acidocaldarius cells developed an electrochemical potential of H+ ions, consisting mainly of a proton gradient and a small inside-negative membrane potential. The steady-state proton motive force of 140 to 160 mV was collapsed by protonophores, while N,N'-dicyclohexylcarbodiimide (DCCD) caused a hyperpolarization of the membrane, as expected for a reagent commonly used to inhibit the flux through proton channels of F0F1-type ATP synthases. Cellular ATP content was strongly related to the proton motive force generated by respiration and declined rapidly, either by uncoupling or by action of DCCD, which in turn induced a marked respiratory control effect. This observation strongly supports the operation of chemiosmotic ATP synthesis with H+ as the coupling ion. The inhibition of ATP synthesis by [14C]DCCD was correlated with covalent reactions with membrane proteins. The extraction of labeled membranes with organic solvents specifically yielded a readily aggregating proteolipid of 6 to 7 kilodaltons apparent molecular mass. Its amino acid composition revealed significant similarity to the proteolipid found in eubacteria, such as Escherichia coli, as an extremely hydrophobic constituent of the F0 proton channel. Moreover, the N-terminal amino acid sequence of the Sulfolobus proteolipid displays a high degree of homology to eubacterial sequences, as well as to one derived from nucleic acid sequencing of another Sulfolobus strain (K. Denda, J. Konishi, T. Oshima, T. Date, and M. Yoshida, J. Biol. Chem. 264:7119-7121, 1989). Despite certain structural similarities between eucaryotic vacuolar ATPases and the F1-analogous ATPase from Sulfolobus sp. described earlier, the results reported here promote the view that the archaebacterial ATP-synthesizing complex functionally belongs to the F0F1 class of ATPases. These may be considered as phylogenetically conserved catalysts of energy transduction present in all kingdoms of organisms.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altendorf K. Purification of the DCCD-reactive protein of the energy-transducing adenosine triphosphatase complex from Escherichia coli. FEBS Lett. 1977 Feb 1;73(2):271–275. doi: 10.1016/0014-5793(77)80997-6. [DOI] [PubMed] [Google Scholar]
- Azzi A., Casey R. P., Nałecz M. J. The effect of N,N'-dicyclohexylcarbodiimide on enzymes of bioenergetic relevance. Biochim Biophys Acta. 1984 Dec 17;768(3-4):209–226. doi: 10.1016/0304-4173(84)90017-x. [DOI] [PubMed] [Google Scholar]
- Bakker E. P. Membrane potential in a potassium transport-negative mutant of Escherichia coli K-12. The distribution of rubidium in the presence of valinomycin indicates a higher potential than that of the tetraphenylphosphonium cation. Biochim Biophys Acta. 1982 Sep 15;681(3):474–483. doi: 10.1016/0005-2728(82)90190-6. [DOI] [PubMed] [Google Scholar]
- Bakker E. P., Randall L. L. The requirement for energy during export of beta-lactamase in Escherichia coli is fulfilled by the total protonmotive force. EMBO J. 1984 Apr;3(4):895–900. doi: 10.1002/j.1460-2075.1984.tb01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman B. J., Bowman E. J. H+-ATPases from mitochondria, plasma membranes, and vacuoles of fungal cells. J Membr Biol. 1986;94(2):83–97. doi: 10.1007/BF01871190. [DOI] [PubMed] [Google Scholar]
- Bowman E. J., Tenney K., Bowman B. J. Isolation of genes encoding the Neurospora vacuolar ATPase. Analysis of vma-1 encoding the 67-kDa subunit reveals homology to other ATPases. J Biol Chem. 1988 Oct 5;263(28):13994–14001. [PubMed] [Google Scholar]
- Brock T. D., Brock K. M., Belly R. T., Weiss R. L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84(1):54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- Burstein C., Tiankova L., Kepes A. Respiratory control in Escherichia coli K 12. Eur J Biochem. 1979 Mar;94(2):387–392. doi: 10.1111/j.1432-1033.1979.tb12905.x. [DOI] [PubMed] [Google Scholar]
- CHANCE B., WILLIAMS G. R. The respiratory chain and oxidative phosphorylation. Adv Enzymol Relat Subj Biochem. 1956;17:65–134. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobley J. G., Cox J. C. Energy conservation in acidophilic bacteria. Microbiol Rev. 1983 Dec;47(4):579–595. doi: 10.1128/mr.47.4.579-595.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A., Stoeckenius W. Photophosphorylation in Halobacterium halobium. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1234–1238. doi: 10.1073/pnas.71.4.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M., Gambacorta A., Nicolaus B., Giardina P., Poerio E., Buonocore V. Glucose metabolism in the extreme thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem J. 1984 Dec 1;224(2):407–414. doi: 10.1042/bj2240407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denda K., Konishi J., Oshima T., Date T., Yoshida M. A gene encoding the proteolipid subunit of Sulfolobus acidocaldarius ATPase complex. J Biol Chem. 1989 May 5;264(13):7119–7121. [PubMed] [Google Scholar]
- Denda K., Konishi J., Oshima T., Date T., Yoshida M. Molecular cloning of the beta-subunit of a possible non-F0F1 type ATP synthase from the acidothermophilic archaebacterium, Sulfolobus acidocaldarius. J Biol Chem. 1988 Nov 25;263(33):17251–17254. [PubMed] [Google Scholar]
- Denda K., Konishi J., Oshima T., Date T., Yoshida M. The membrane-associated ATPase from Sulfolobus acidocaldarius is distantly related to F1-ATPase as assessed from the primary structure of its alpha-subunit. J Biol Chem. 1988 May 5;263(13):6012–6015. [PubMed] [Google Scholar]
- Fillingame R. H. Purification of the carbodiimide-reactive protein component of the ATP energy-transducing system of Escherichia coli. J Biol Chem. 1976 Nov 10;251(21):6630–6637. [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):2009–2015. [PubMed] [Google Scholar]
- Goulbourne E., Jr, Matin M., Zychlinsky E., Matin A. Mechanism of delta pH maintenance in active and inactive cells of an obligately acidophilic bacterium. J Bacteriol. 1986 Apr;166(1):59–65. doi: 10.1128/jb.166.1.59-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T., Sebald W. The dicyclohexylcarbodiimide-binding protein of the mitochondrial ATPase complex from beef heart. Isolation and amino acid composition. FEBS Lett. 1978 Oct 15;94(2):218–222. doi: 10.1016/0014-5793(78)80941-7. [DOI] [PubMed] [Google Scholar]
- Hartl T., Grossebüter W., Görisch H., Stezowski J. J. Crystalline NAD/NADP-dependent malate dehydrogenase; the enzyme from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biol Chem Hoppe Seyler. 1987 Mar;368(3):259–267. doi: 10.1515/bchm3.1987.368.1.259. [DOI] [PubMed] [Google Scholar]
- Hartmann R., Sickinger H. D., Oesterhelt D. Anaerobic growth of halobacteria. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3821–3825. doi: 10.1073/pnas.77.7.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein L. I., Kristjansson H., Altekar W. The purification and subunit structure of a membrane-bound ATPase from the Archaebacterium Halobacterium saccharovorum. Biochem Biophys Res Commun. 1987 Aug 31;147(1):295–300. doi: 10.1016/s0006-291x(87)80120-1. [DOI] [PubMed] [Google Scholar]
- Hoppe J., Sebald W. Amino acid sequence of the proteolipid subunit of the proton-translocating ATPase complex from the thermophilic bacterium PS-3. Eur J Biochem. 1980;107(1):57–65. doi: 10.1111/j.1432-1033.1980.tb04624.x. [DOI] [PubMed] [Google Scholar]
- Hoppe J., Sebald W. The proton conducting F0-part of bacterial ATP synthases. Biochim Biophys Acta. 1984 Apr 9;768(1):1–27. doi: 10.1016/0304-4173(84)90005-3. [DOI] [PubMed] [Google Scholar]
- Hsung J. C., Haug A. Membrane potential of Thermoplasma acidophila. FEBS Lett. 1977 Jan 15;73(1):47–50. doi: 10.1016/0014-5793(77)80011-2. [DOI] [PubMed] [Google Scholar]
- Inatomi K. Characterization and purification of the membrane-bound ATPase of the archaebacterium Methanosarcina barkeri. J Bacteriol. 1986 Sep;167(3):837–841. doi: 10.1128/jb.167.3.837-841.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashket E. R. The proton motive force in bacteria: a critical assessment of methods. Annu Rev Microbiol. 1985;39:219–242. doi: 10.1146/annurev.mi.39.100185.001251. [DOI] [PubMed] [Google Scholar]
- Konishi J., Wakagi T., Oshima T., Yoshida M. Purification and properties of the ATPase solubilized from membranes of an acidothermophilic archaebacterium, Sulfolobus acidocaldarius. J Biochem. 1987 Dec;102(6):1379–1387. doi: 10.1093/oxfordjournals.jbchem.a122184. [DOI] [PubMed] [Google Scholar]
- Kopecký J., Houstek J., Szarska E., Drahota Z. Electrophoretic behavior of the H+-ATPase proteolipid from bovine heart mitochondria. J Bioenerg Biomembr. 1986 Dec;18(6):507–519. doi: 10.1007/BF00743147. [DOI] [PubMed] [Google Scholar]
- Krulwich T. A., Davidson L. F., Filip S. J., Jr, Zuckerman R. S., Guffanti A. A. The protonmotive force and beta-galactoside transport in Bacillus acidocaldarius. J Biol Chem. 1978 Jul 10;253(13):4599–4603. [PubMed] [Google Scholar]
- Krulwich T. A., Guffanti A. A. Regulation of internal pH in acidophilic and alkalophilic bacteria. Methods Enzymol. 1986;125:352–365. doi: 10.1016/s0076-6879(86)25030-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lübben M., Lünsdorf H., Schäfer G. Archaebacterial ATPase: studies on subunit composition and quaternary structure of the F1-analogous ATPase from Sulfolobus acidocaldarius. Biol Chem Hoppe Seyler. 1988 Nov;369(11):1259–1266. doi: 10.1515/bchm3.1988.369.2.1259. [DOI] [PubMed] [Google Scholar]
- Lübben M., Lünsdorf H., Schäfer G. The plasma membrane ATPase of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Purification and immunological relationships to F1-ATPases. Eur J Biochem. 1987 Sep 1;167(2):211–219. doi: 10.1111/j.1432-1033.1987.tb13325.x. [DOI] [PubMed] [Google Scholar]
- Lübben M., Schäfer G. A plasma-membrane associated ATPase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Eur J Biochem. 1987 May 4;164(3):533–540. doi: 10.1111/j.1432-1033.1987.tb11159.x. [DOI] [PubMed] [Google Scholar]
- Mandel M., Moriyama Y., Hulmes J. D., Pan Y. C., Nelson H., Nelson N. cDNA sequence encoding the 16-kDa proteolipid of chromaffin granules implies gene duplication in the evolution of H+-ATPases. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5521–5524. doi: 10.1073/pnas.85.15.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin A., Wilson B., Zychlinsky E., Matin M. Proton motive force and the physiological basis of delta pH maintenance in thiobacillus acidophilus. J Bacteriol. 1982 May;150(2):582–591. doi: 10.1128/jb.150.2.582-591.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels M., Bakker E. P. Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J Bacteriol. 1985 Jan;161(1):231–237. doi: 10.1128/jb.161.1.231-237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukohata Y., Ihara K., Yoshida M., Konishi J., Sugiyama Y., Yoshida M. The halobacterial H+-translocating ATP synthase relates to the eukaryotic anion-sensitive H+-ATPase. Arch Biochem Biophys. 1987 Dec;259(2):650–653. doi: 10.1016/0003-9861(87)90532-7. [DOI] [PubMed] [Google Scholar]
- Nanba T., Mukohata Y. A membrane-bound ATPase from Halobacterium halobium: purification and characterization. J Biochem. 1987 Sep;102(3):591–598. doi: 10.1093/oxfordjournals.jbchem.a122092. [DOI] [PubMed] [Google Scholar]
- Nelson N., Taiz L. The evolution of H+-ATPases. Trends Biochem Sci. 1989 Mar;14(3):113–116. doi: 10.1016/0968-0004(89)90134-5. [DOI] [PubMed] [Google Scholar]
- Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977 Dec;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Pressman B. C. Biological applications of ionophores. Annu Rev Biochem. 1976;45:501–530. doi: 10.1146/annurev.bi.45.070176.002441. [DOI] [PubMed] [Google Scholar]
- Rottenberg H. The measurement of membrane potential and deltapH in cells, organelles, and vesicles. Methods Enzymol. 1979;55:547–569. doi: 10.1016/0076-6879(79)55066-6. [DOI] [PubMed] [Google Scholar]
- Schneider E., Altendorf K. All three subunits are required for the reconstitution of an active proton channel (F0) of Escherichia coli ATP synthase (F1F0). EMBO J. 1985 Feb;4(2):515–518. doi: 10.1002/j.1460-2075.1985.tb03658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Altendorf K. Bacterial adenosine 5'-triphosphate synthase (F1F0): purification and reconstitution of F0 complexes and biochemical and functional characterization of their subunits. Microbiol Rev. 1987 Dec;51(4):477–497. doi: 10.1128/mr.51.4.477-497.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Searcy D. G. Thermoplasma acidophilum: intracellular pH and potassium concentration. Biochim Biophys Acta. 1976 Nov 18;451(1):278–286. doi: 10.1016/0304-4165(76)90278-6. [DOI] [PubMed] [Google Scholar]
- Sebald W., Graf T., Lukins H. B. The dicyclohexylcarbodiimide-binding protein of the mitochondrial ATPase complex from Neurospora crassa and Saccharomyces cerevisiae. Identification and isolation. Eur J Biochem. 1979 Feb 1;93(3):587–599. doi: 10.1111/j.1432-1033.1979.tb12859.x. [DOI] [PubMed] [Google Scholar]
- Senior A. E. ATP synthesis by oxidative phosphorylation. Physiol Rev. 1988 Jan;68(1):177–231. doi: 10.1152/physrev.1988.68.1.177. [DOI] [PubMed] [Google Scholar]
- Steffens K., Di Gioia A., Deckers-Hebestreit G., Altendorf K. Structural and functional relationship of ATP synthases (F1F0) from Escherichia coli and the thermophilic bacterium PS3. J Biol Chem. 1987 May 5;262(13):6334–6338. [PubMed] [Google Scholar]
- Wakao H., Wakagi T., Oshima T. Purification and properties of NADH dehydrogenase from a thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. J Biochem. 1987 Aug;102(2):255–262. doi: 10.1093/oxfordjournals.jbchem.a122049. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Fearnley I. M., Gay N. J., Gibson B. W., Northrop F. D., Powell S. J., Runswick M. J., Saraste M., Tybulewicz V. L. Primary structure and subunit stoichiometry of F1-ATPase from bovine mitochondria. J Mol Biol. 1985 Aug 20;184(4):677–701. doi: 10.1016/0022-2836(85)90313-4. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- Watters C. A one-step biuret assay for protein in the presence of detergent. Anal Biochem. 1978 Aug 1;88(2):695–698. doi: 10.1016/0003-2697(78)90475-x. [DOI] [PubMed] [Google Scholar]
- Zaritsky A., Kihara M., Macnab R. M. Measurement of membrane potential in Bacillus subtilis: a comparison of lipophilic cations, rubidium ion, and a cyanine dye as probes. J Membr Biol. 1981;63(3):215–231. doi: 10.1007/BF01870983. [DOI] [PubMed] [Google Scholar]
- Zimniak L., Dittrich P., Gogarten J. P., Kibak H., Taiz L. The cDNA sequence of the 69-kDa subunit of the carrot vacuolar H+-ATPase. Homology to the beta-chain of F0F1-ATPases. J Biol Chem. 1988 Jul 5;263(19):9102–9112. [PubMed] [Google Scholar]