Abstract
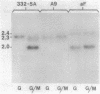
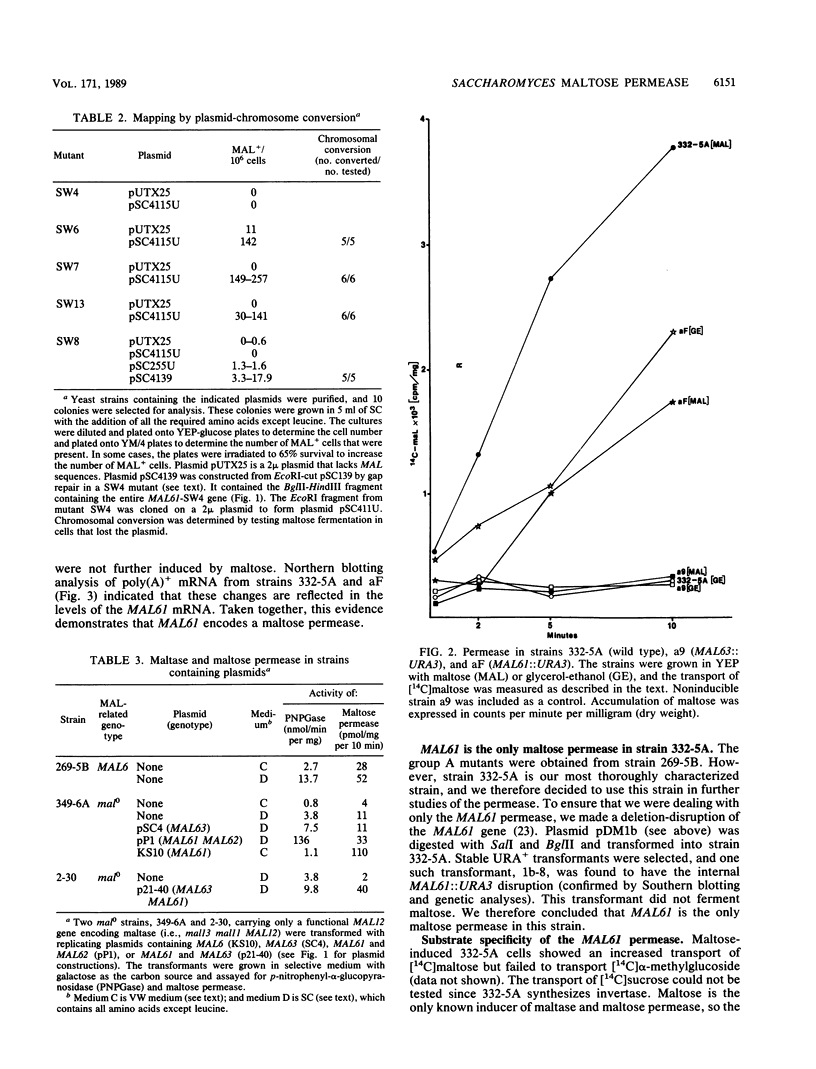
Saccharomyces yeasts ferment several alpha-glucosides including maltose, maltotriose, turanose, alpha-methylglucoside, and melezitose. In the utilization of these sugars transport is the rate-limiting step. Several groups of investigators have described the characteristics of the maltose permease (D. E. Kroon and V. V. Koningsberger, Biochim. Biophys. Acta 204:590-609, 1970; R. Serrano, Eur. J. Biochem. 80:97-102, 1977). However, Saccharomyces contains multiple alpha-glucoside transport systems, and these studies have never been performed on a genetically defined strain shown to have only a single permease gene. In this study we isolated maltose-negative mutants in a MAL6 strain and, using a high-resolution mapping technique, we showed that one class of these mutants, the group A mutants, mapped to the MAL61 gene (a member of the MAL6 gene complex). An insertion into the N-terminal-coding region of MAL61 resulted in the constitutive production of MAL61 mRNA and rendered the maltose permease similarly constitutive. Transformation by high-copy-number plasmids containing the MAL61 gene also led to an increase in the maltose permease. A deletion-disruption of MAL61 completely abolished maltose transport activity. Taken together, these results prove that this strain has only a single maltose permease and that this permease is the product of the MAL61 gene. This permease is able to transport maltose and turanose but cannot transport maltotriose, alpha-methylglucoside, or melezitose. The construction of strains with only a single permease will allow us to identify other maltose-inducible transport systems by simple genetic tests and should lead to the identification and characterization of the multiple genes and gene products involved in alpha-glucoside transport in Saccharomyces yeasts.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang Y. S., Dubin R. A., Perkins E., Forrest D., Michels C. A., Needleman R. B. MAL63 codes for a positive regulator of maltose fermentation in Saccharomyces cerevisiae. Curr Genet. 1988 Sep;14(3):201–209. doi: 10.1007/BF00376740. [DOI] [PubMed] [Google Scholar]
- Charron M. J., Dubin R. A., Michels C. A. Structural and functional analysis of the MAL1 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3891–3899. doi: 10.1128/mcb.6.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron M. J., Michels C. A. The naturally occurring alleles of MAL1 in Saccharomyces species evolved by various mutagenic processes including chromosomal rearrangement. Genetics. 1988 Sep;120(1):83–93. doi: 10.1093/genetics/120.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. D., Goldenthal M. J., Buchferer B., Marmur J. Mutational analysis of the MAL1 locus of Saccharomyces: identification and functional characterization of three genes. Mol Gen Genet. 1984;196(2):208–216. doi: 10.1007/BF00328052. [DOI] [PubMed] [Google Scholar]
- Cohen J. D., Goldenthal M. J., Chow T., Buchferer B., Marmur J. Organization of the MAL loci of Saccharomyces. Physical identification and functional characterization of three genes at the MAL6 locus. Mol Gen Genet. 1985;200(1):1–8. doi: 10.1007/BF00383304. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Dubin R. A., Needleman R. B., Gossett D., Michels C. A. Identification of the structural gene encoding maltase within the MAL6 locus of Saccharomyces carlsbergensis. J Bacteriol. 1985 Nov;164(2):605–610. doi: 10.1128/jb.164.2.605-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin R. A., Perkins E. L., Needleman R. B., Michels C. A. Identification of a second trans-acting gene controlling maltose fermentation in Saccharomyces carlsbergensis. Mol Cell Biol. 1986 Aug;6(8):2757–2765. doi: 10.1128/mcb.6.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Rose M., Botstein D. Homologous Recombination between Episomal Plasmids and Chromosomes in Yeast. Genetics. 1983 Dec;105(4):843–856. doi: 10.1093/genetics/105.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. H., Marmur J. Primary structure of the maltase gene of the MAL6 locus of Saccharomyces carlsbergensis. Gene. 1986;41(1):75–84. doi: 10.1016/0378-1119(86)90269-6. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotyk A., Michaljanicová D. Uptake of trehalose by Saccharomyces cerevisiae. J Gen Microbiol. 1979 Feb;110(2):323–332. doi: 10.1099/00221287-110-2-323. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. Fine structure mapping in yeast with sunlamp radiation. Genetics. 1974 Apr;76(4):723–733. doi: 10.1093/genetics/76.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels C. A., Needleman R. B. The dispersed, repeated family of MAL loci in Saccharomyces spp. J Bacteriol. 1984 Mar;157(3):949–952. doi: 10.1128/jb.157.3.949-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman R. B., Kaback D. B., Dubin R. A., Perkins E. L., Rosenberg N. G., Sutherland K. A., Forrest D. B., Michels C. A. MAL6 of Saccharomyces: a complex genetic locus containing three genes required for maltose fermentation. Proc Natl Acad Sci U S A. 1984 May;81(9):2811–2815. doi: 10.1073/pnas.81.9.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman R., Eaton N. R. Selection of yeast mutants constitutive for maltase synthesis. Mol Gen Genet. 1974;133(2):135–140. doi: 10.1007/BF00264834. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Serrano R. Energy requirements for maltose transport in yeast. Eur J Biochem. 1977 Oct 17;80(1):97–102. doi: 10.1111/j.1432-1033.1977.tb11861.x. [DOI] [PubMed] [Google Scholar]
- de Kroon R. A., Koningsberger V. V. An inducible transport system for alpha-glucosides in protoplasts of Saccharomyces carlsbergensis. Biochim Biophys Acta. 1970 Apr 15;204(2):590–609. doi: 10.1016/0005-2787(70)90178-4. [DOI] [PubMed] [Google Scholar]
- ten Berge A. M., Zoutewelle G., van de Poll K. W. Regulation of maltose fermentation in Saccharomyces carlsbergensis. I. The function of the gene MAL6, as recognized by mal6-mutants. Mol Gen Genet. 1973 Jul 2;123(3):233–246. doi: 10.1007/BF00271242. [DOI] [PubMed] [Google Scholar]