Abstract
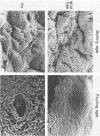
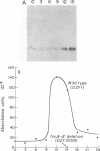
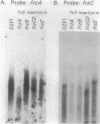
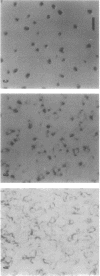
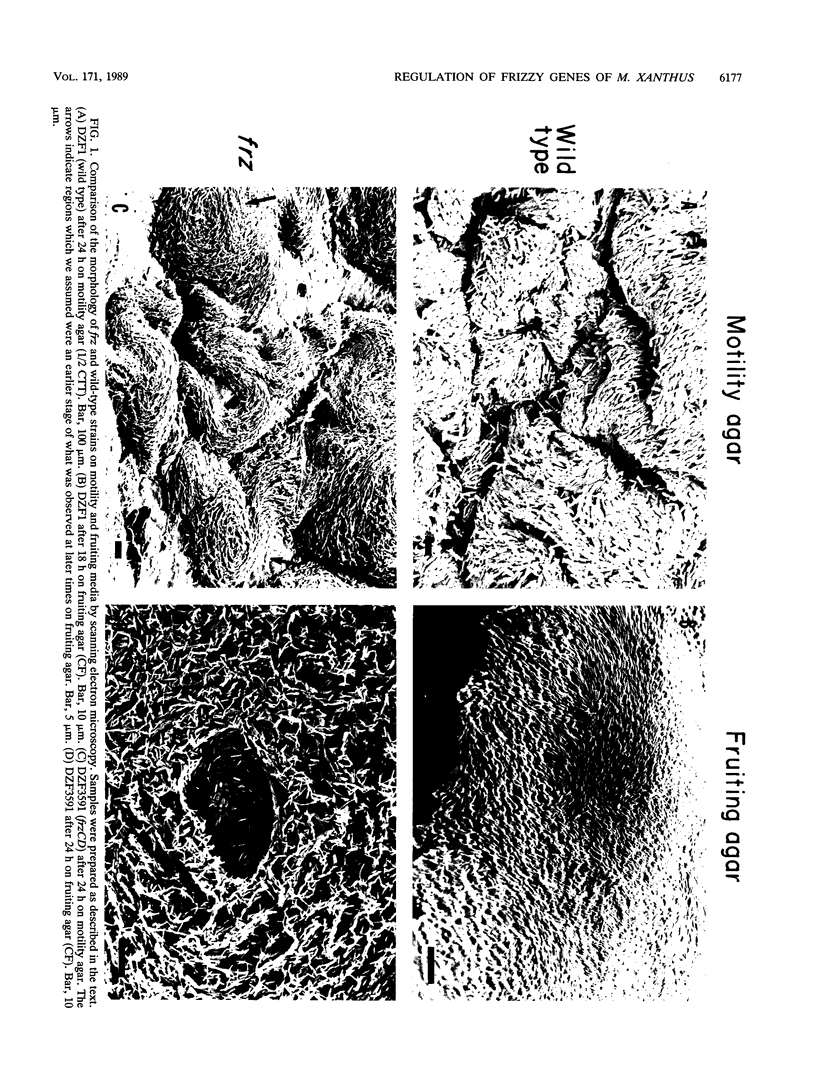
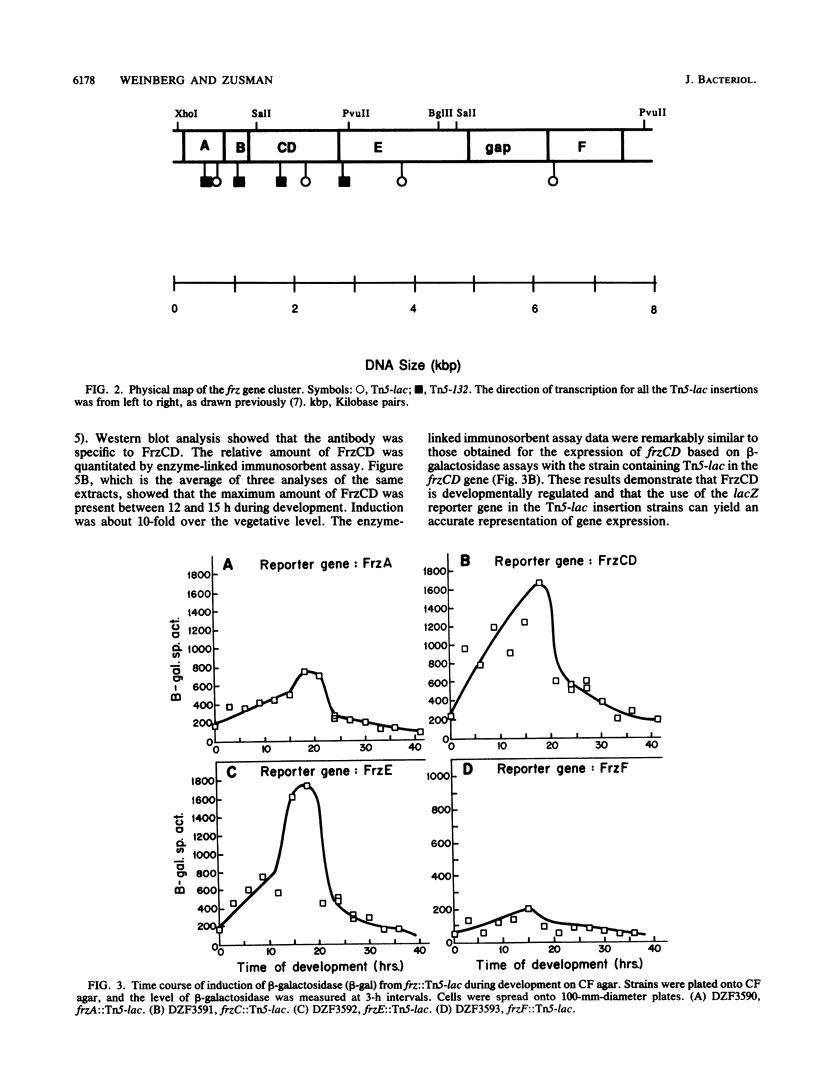
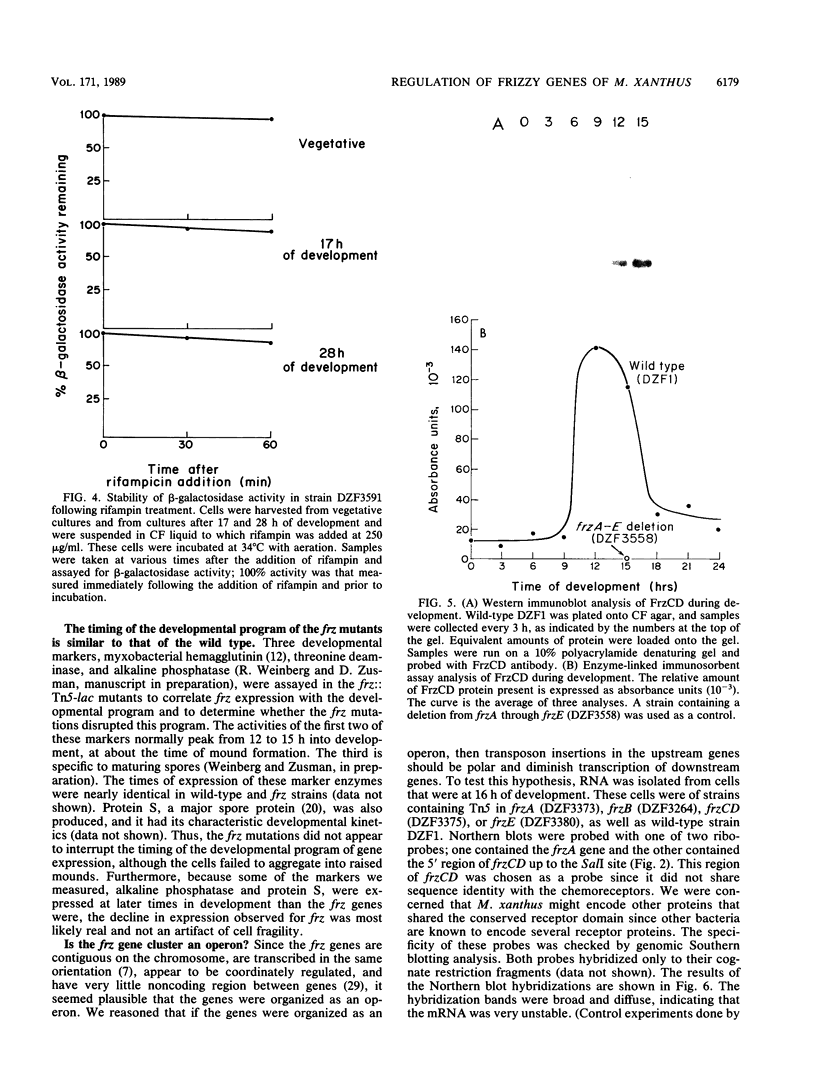
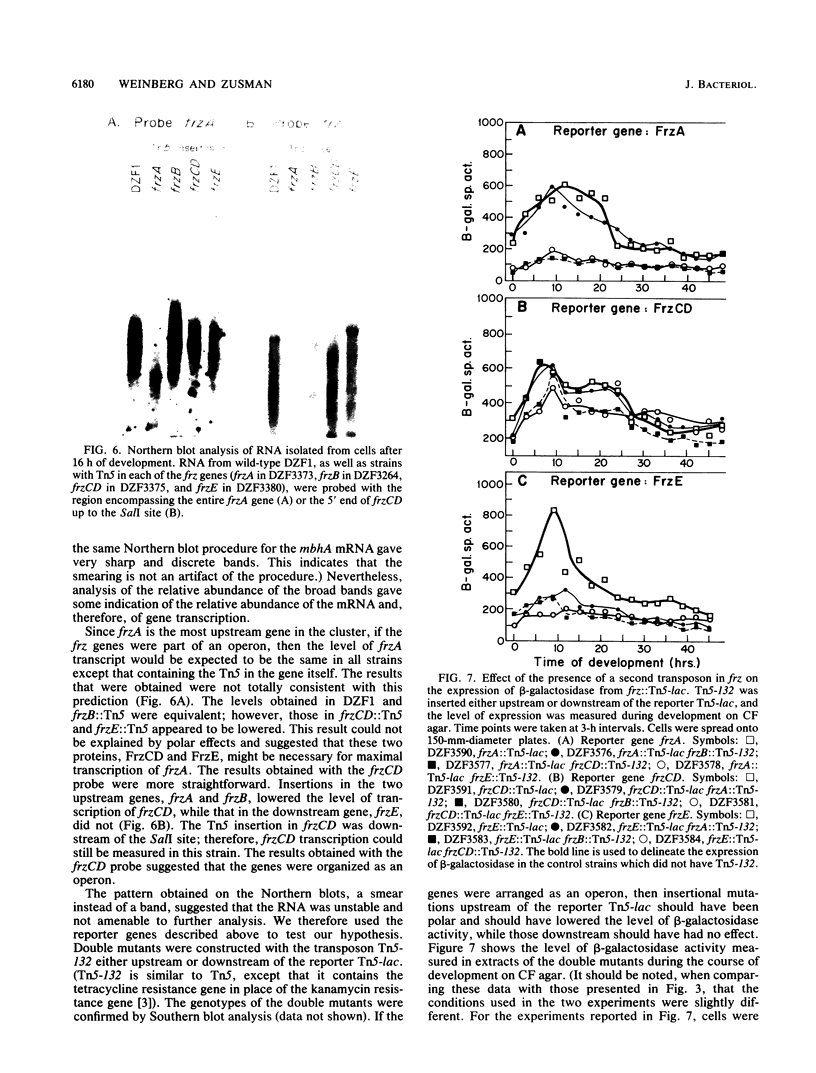
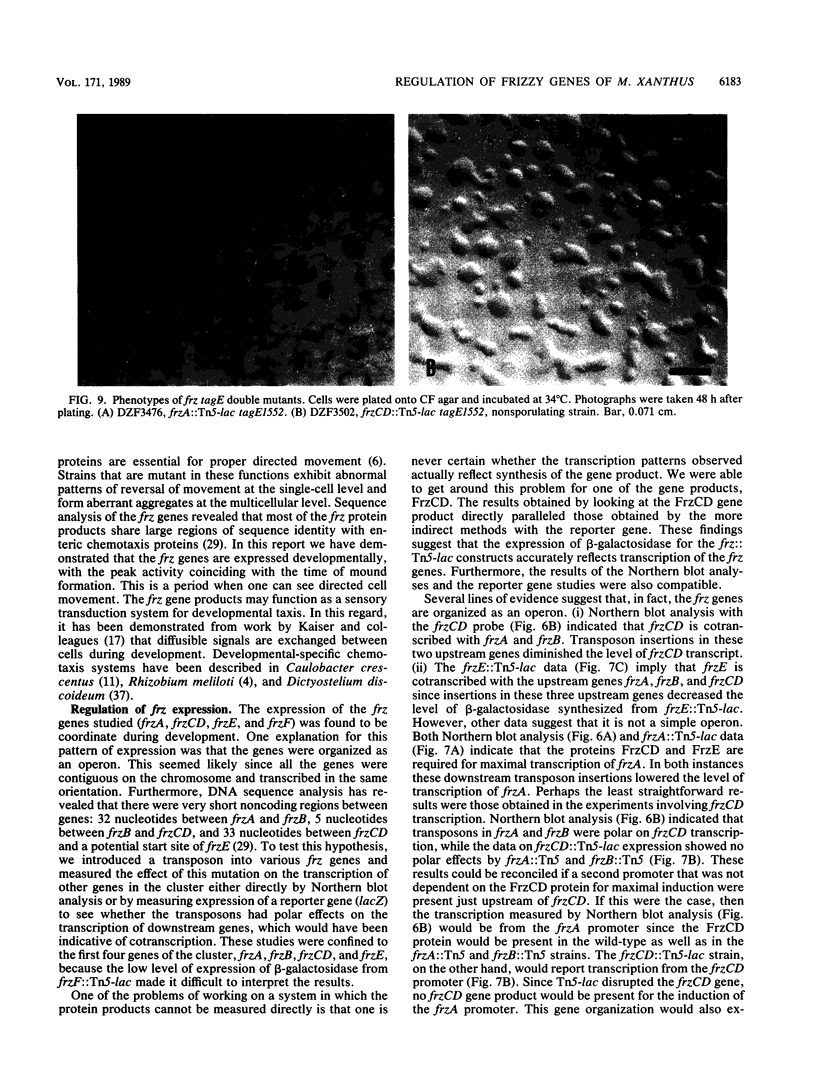
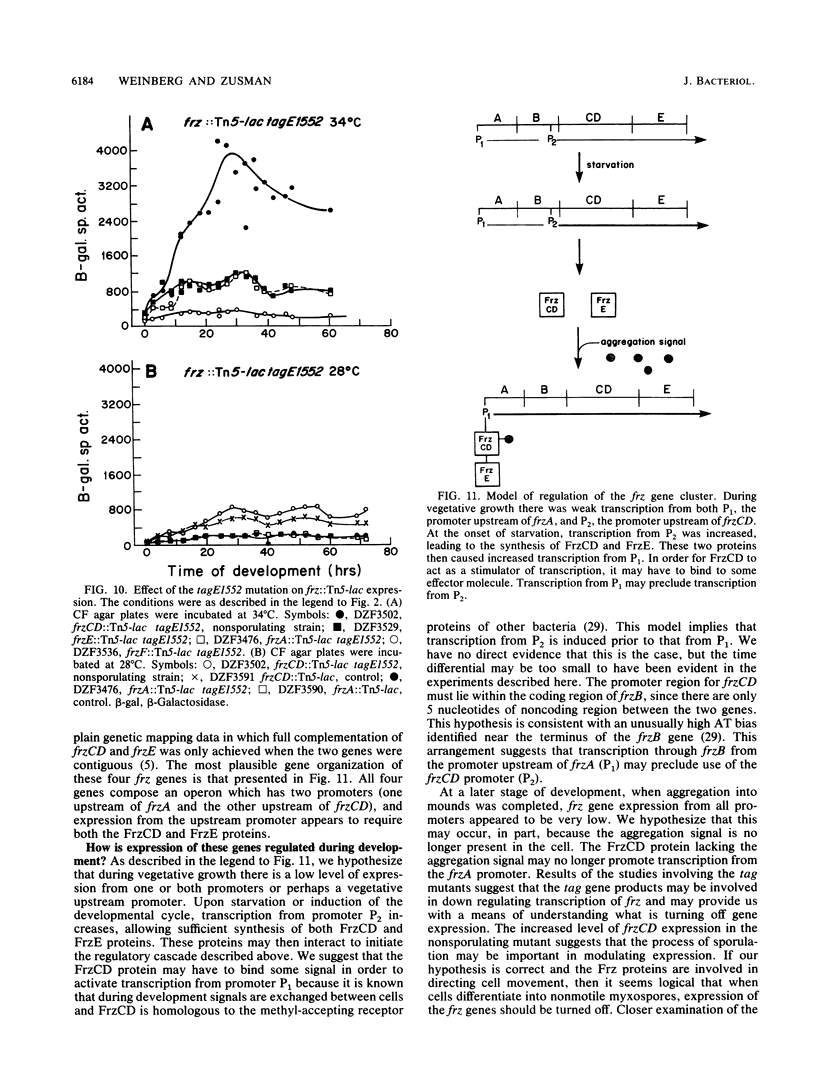
The frizzy (frz) mutants of Myxococcus xanthus are unable to form fruiting bodies. Instead of forming discrete mounds, these strains aggregate as filaments which have a circular and tangled appearance. Mutations leading to this phenotype have been mapped to five complementation groups, frzA, frzB, frzCD, frzE, and frzF. All have been found to be involved in the control of directional movement of the bacteria and, except for frzB, to be homologous to the chemotaxis genes of enteric bacteria. In this report we present a study of the regulation of expression of the first four genes of the frz gene cluster (frzA, frzB, frzCD, and frzE) by using Tn5-lac transcriptional fusions as reporters of gene expression. We found that these frz genes are developmentally regulated, with their transcription peaking at about the time of early mound formation (12 to 18 h). Analysis of FrzCD expression by enzyme-linked immunosorbent assay showed a 10-fold greater induction at 15 h of development over the level of vegetative cell expression. Northern blot hybridization analysis suggested that the frz genes were arranged as an operon. To test this hypothesis, double mutants were constructed which contained Tn5-132 either upstream or downstream of the reporter Tn5-lac. The expression of the frz genes in the double mutants was consistent with the hypothesis that the first four genes (frzA, frzB, frzCD, and frzE) are organized as an operon with an internal promoter. Insertion mutations in frzCD lowered gene expression whether they were upstream or downstream of the reporter Tn5-lac, suggesting that the FrzCD protein regulates transcription of the entire operon from a promoter upstream of frzA. Evidence is presented suggesting that FrzE is required for induction of transcription as well. When frz mutations were placed in strains that were unable to aggregate (tag), the frz genes were expressed at an elevated level on fruiting agar; this high level of expression was maintained for several days. These results suggest that the tag gene products interact with the frz functions.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnosti D. N., Chamberlin M. J. Secondary sigma factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Feb;86(3):830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Weiss A., Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980 May;142(2):439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman K., Gulash-Hoffee M., Hovestadt R. E., Larosiliere R. C., Ronco P. G., 2nd, Su L. Physiology of behavioral mutants of Rhizobium meliloti: evidence for a dual chemotaxis pathway. J Bacteriol. 1988 Jul;170(7):3249–3254. doi: 10.1128/jb.170.7.3249-3254.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. "Frizzy" genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. Analysis of the products of the Myxococcus xanthus frz genes. J Bacteriol. 1986 May;166(2):673–678. doi: 10.1128/jb.166.2.673-678.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D., Zusman D. R. Cloning and complementation analysis of the "Frizzy" genes of Myxococcus xanthus. Mol Gen Genet. 1985;198(2):243–254. doi: 10.1007/BF00383002. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bryan R., Champer R., Gomes S., Ely B., Shapiro L. Separation of temporal control and trans-acting modulation of flagellin and chemotaxis genes in Caulobacter. Mol Gen Genet. 1987 Feb;206(2):300–306. doi: 10.1007/BF00333588. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Champer R., Dingwall A., Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987 Mar 5;194(1):71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- Cumsky M., Zusman D. R. Myxobacterial hemagglutinin: a development-specific lectin of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5505–5509. doi: 10.1073/pnas.76.11.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DWORKIN M., GIBSON S. M. A SYSTEM FOR STUDYING MICROBIAL MORPHOGENESIS: RAPID FORMATION OF MICROCYSTS IN MYXOCOCCUS XANTHUS. Science. 1964 Oct 9;146(3641):243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
- Garriga G., Bertrand H., Lambowitz A. M. RNA splicing in Neurospora mitochondria: nuclear mutants defective in both splicing and 3' end synthesis of the large rRNA. Cell. 1984 Mar;36(3):623–634. doi: 10.1016/0092-8674(84)90342-8. [DOI] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell. 1984 Dec;39(3 Pt 2):631–641. doi: 10.1016/0092-8674(84)90470-7. [DOI] [PubMed] [Google Scholar]
- Gitt M. A., Wang L. F., Doi R. H. A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J Biol Chem. 1985 Jun 25;260(12):7178–7185. [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Márquez L. M., Chamberlin M. J. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J Bacteriol. 1988 Apr;170(4):1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska-Grynkiewicz K., Kustu S. Evidence that nitrogen regulatory gene ntrC of Salmonella typhimurium is transcribed from the glnA promoter as well as from a separate ntr promoter. Mol Gen Genet. 1984;193(1):135–142. doi: 10.1007/BF00327426. [DOI] [PubMed] [Google Scholar]
- Kreuzer K., Pratt C., Torriani A. Genetic analysis of regulatory mutants of alkaline phosphatase of E. coli. Genetics. 1975 Nov;81(3):459–468. doi: 10.1093/genetics/81.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. J., Weinberg R. A., Zusman D. R. "Frizzy" aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci U S A. 1989 Jan;86(2):424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983 Jul;155(1):317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Reexamination of the role of autolysis in the development of Myxococcus xanthus. J Bacteriol. 1988 Sep;170(9):4103–4112. doi: 10.1128/jb.170.9.4103-4112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R., Talbert P. B., Houts S. E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983 Jul;155(1):265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J. E., Fisher P. R., Gerisch G. Selection of chemotaxis mutants of Dictyostelium discoideum. J Cell Biol. 1987 Jan;104(1):151–161. doi: 10.1083/jcb.104.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman D. R. "Frizzy" mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982 Jun;150(3):1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]