Abstract
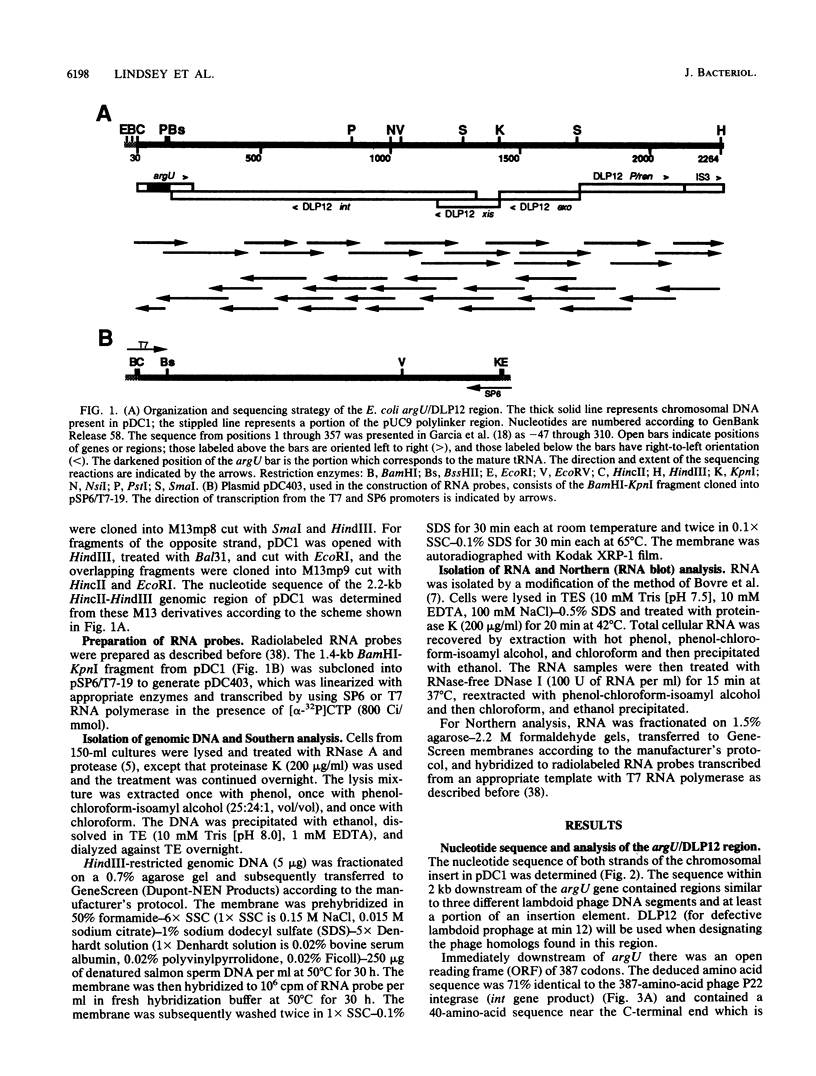
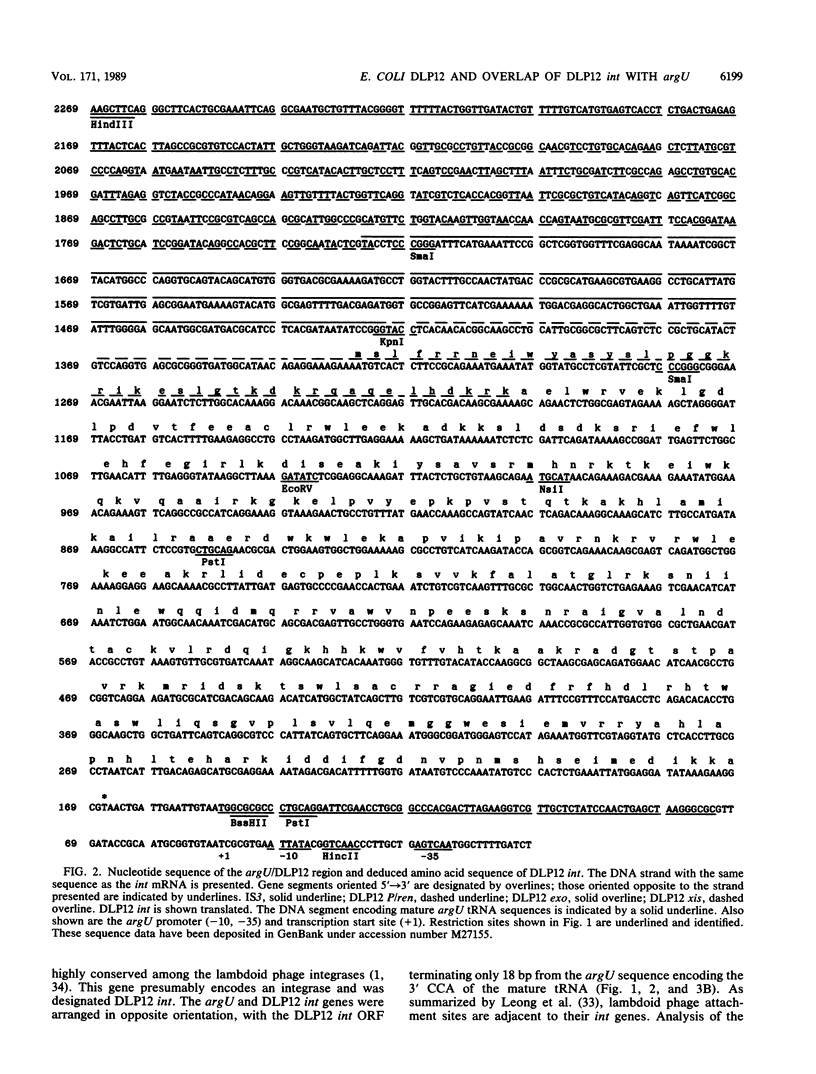
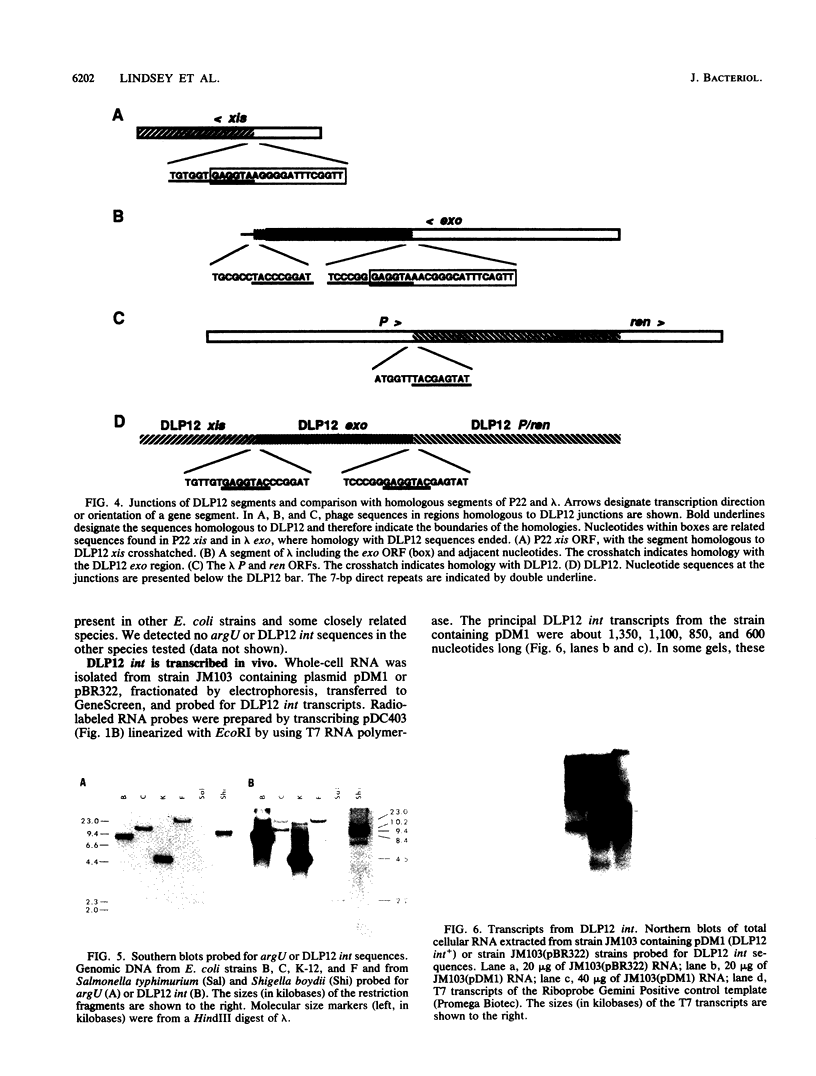
The argU (dnaY) gene of Escherichia coli is located, in clockwise orientation, at 577.5 kilobases (kb) on the chromosome physical map. There was a cryptic prophage spanning the 2 kb immediately downstream of argU that consisted of sequences similar to the phage P22 int gene, a portion of the P22 xis gene, and portions of the exo, P, and ren genes of bacteriophage lambda. This cryptic prophage was designated DLP12, for defective lambdoid prophage at 12 min. Immediately clockwise of DLP12 was the IS3 alpha 4 beta 4 insertion element. The argU and DLP12 int genes overlapped at their 3' ends, and argU contained sequence homologous to a portion of the phage P22 attP site. Additional homologies to lambdoid phages were found in the 25 kb clockwise of argU. These included the cryptic prophage qsr' (P. J. Highton, Y. Chang, W. R. Marcotte, Jr., and C. A. Schnaitman, J. Bacteriol. 162:256-262, 1985), a sequence homologous to a portion of lambda orf-194, and an attR homolog. Inasmuch as the DLP12 att int xis exo P/ren region, the qsr' region, and homologs of orf-194 and attR were arranged in the same order and orientation as the lambdoid prophage counterparts, we propose that the designation DLP12 be applied to all these sequences. This organization of the DLP12 sequences and the presence of the argU/DLP12 int pair in several E. coli strains and closely related species suggest that DLP12 might be an ancestral lambdoid prophage. Moreover, the presence of similar sequences at the junctions of DLP12 segments and their phage counterparts suggests that a common mechanism could have transferred these DLP12 segments to more recent phages.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T., Nielsen A., Hansen F. G. Isolation, characterization, and nucleotide sequence of appY, a regulatory gene for growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1989 Mar;171(3):1683–1691. doi: 10.1128/jb.171.3.1683-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasband A. J., Marcotte W. R., Jr, Schnaitman C. A. Structure of the lc and nmpC outer membrane porin protein genes of lambdoid bacteriophage. J Biol Chem. 1986 Sep 25;261(27):12723–12732. [PubMed] [Google Scholar]
- Blum P., Holzschu D., Kwan H. S., Riggs D., Artz S. Gene replacement and retrieval with recombinant M13mp bacteriophages. J Bacteriol. 1989 Jan;171(1):538–546. doi: 10.1128/jb.171.1.538-546.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Dalrymple B., Mattick J. S. Genes encoding threonine tRNAs with the anticodon CGU from Escherichia coli and Pseudomonas aeruginosa. Biochem Int. 1986 Oct;13(4):547–553. [PubMed] [Google Scholar]
- Deonier R. C., Oh G. R., Hu M. Further mapping of IS2 and IS3 in the lac-purE region of the Escherichia coli K-12 genome: structure of the F-prime ORF203. J Bacteriol. 1977 Feb;129(2):1129–1140. doi: 10.1128/jb.129.2.1129-1140.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deonier R. C., Yun K., Kuppermann M. Gamma delta-mediated deletions of chromosomal segments on F-prime plasmids. Mol Gen Genet. 1983;190(1):42–50. doi: 10.1007/BF00330322. [DOI] [PubMed] [Google Scholar]
- Deutch A. H., Rushlow K. E., Smith C. J. Analysis of the Escherichia coli proBA locus by DNA and protein sequencing. Nucleic Acids Res. 1984 Aug 10;12(15):6337–6355. doi: 10.1093/nar/12.15.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M. J., Ozeki H. Structure and organization of the transfer ribonucleic acid genes of Escherichia coli K-12. Microbiol Rev. 1985 Dec;49(4):379–397. doi: 10.1128/mr.49.4.379-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. M., Mar P. K., Mullin D. A., Walker J. R., Prather N. E. The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986 May 9;45(3):453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- Gayda R. C., Avni H., Berg P. E., Markovitz A. Outer membrane protein a and other polypeptides regulate capsular polysaccharide synthesis in E. coli K-12. Mol Gen Genet. 1979 Oct 1;175(3):325–332. doi: 10.1007/BF00397232. [DOI] [PubMed] [Google Scholar]
- Gordon G., Gayda R. C., Markovitz A. Sequence of the regulatory region of omp T, the gene specifying major outer membrane protein a (3b) of Escherichia coli K-12: implications for regulation and processing. Mol Gen Genet. 1984;193(3):414–421. doi: 10.1007/BF00382077. [DOI] [PubMed] [Google Scholar]
- Grodberg J., Lundrigan M. D., Toledo D. L., Mangel W. F., Dunn J. J. Complete nucleotide sequence and deduced amino acid sequence of the ompT gene of Escherichia coli K-12. Nucleic Acids Res. 1988 Feb 11;16(3):1209–1209. doi: 10.1093/nar/16.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley R. G., Hu M., Timmons M., Yun K., Deonier R. C. A partial restriction map of the proA-purE region of the Escherichia coli K12 chromosome. Gene. 1983 May-Jun;22(2-3):281–287. doi: 10.1016/0378-1119(83)90113-0. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J. M., Chu H., Irwin C. A., Walker J. R. Isolation and characterization of dnaX and dnaY temperature-sensitive mutants of Escherichia coli. Genetics. 1979 Aug;92(4):1041–1059. doi: 10.1093/genetics/92.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highton P. J., Chang Y., Marcotte W. R., Jr, Schnaitman C. A. Evidence that the outer membrane protein gene nmpC of Escherichia coli K-12 lies within the defective qsr' prophage. J Bacteriol. 1985 Apr;162(1):256–262. doi: 10.1128/jb.162.1.256-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe I., Roth J. Specialized transducing phages derived from salmonella phage P22. Genetics. 1974 Apr;76(4):633–654. doi: 10.1093/genetics/76.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Ohtsubo E., Davidson N. Electron microscopic heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F13 and related F-primes. J Bacteriol. 1975 May;122(2):749–763. doi: 10.1128/jb.122.2.749-763.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Jasin M., Schimmel P. Deletion of an essential gene in Escherichia coli by site-specific recombination with linear DNA fragments. J Bacteriol. 1984 Aug;159(2):783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp E. H., Minton N. P., Mann N. H. Complete nucleotide sequence and deduced amino acid sequence of the M5 polypeptide gene of Escherichia coli. Nucleic Acids Res. 1987 May 11;15(9):3924–3924. doi: 10.1093/nar/15.9.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kushner S. R., Nagaishi H., Templin A., Clark A. J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci U S A. 1971 Apr;68(4):824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S. E., Oser A. B., Lesser C. F., Youderian P., Susskind M. M., Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986 Jun 20;189(4):603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- Lundrigan M. D., Earhart C. F. Gene envY of Escherichia coli K-12 affects thermoregulation of major porin expression. J Bacteriol. 1984 Jan;157(1):262–268. doi: 10.1128/jb.157.1.262-268.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundrigan M. D., Friedrich M. J., Kadner R. J. Nucleotide sequence of the Escherichia coli porin thermoregulatory gene envY. Nucleic Acids Res. 1989 Jan 25;17(2):800–800. doi: 10.1093/nar/17.2.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin D. A., Garcia G. M., Walker J. R. An E. coli DNA fragment 118 base pairs in length provides dnaY+ complementing activity. Cell. 1984 Jun;37(2):669–674. doi: 10.1016/0092-8674(84)90399-4. [DOI] [PubMed] [Google Scholar]
- Pasloske B. L., Finlay B. B., Paranchych W. Cloning and sequencing of the Pseudomonas aeruginosa PAK pilin gene. FEBS Lett. 1985 Apr 22;183(2):408–412. doi: 10.1016/0014-5793(85)80821-8. [DOI] [PubMed] [Google Scholar]
- Pierson L. S., 3rd, Kahn M. L. Integration of satellite bacteriophage P4 in Escherichia coli. DNA sequences of the phage and host regions involved in site-specific recombination. J Mol Biol. 1987 Aug 5;196(3):487–496. doi: 10.1016/0022-2836(87)90026-x. [DOI] [PubMed] [Google Scholar]
- Redfield R. J., Campbell A. M. Structure of cryptic lambda prophages. J Mol Biol. 1987 Dec 5;198(3):393–404. doi: 10.1016/0022-2836(87)90289-0. [DOI] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989 Mar 11;17(5):1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht K. R., Gordon G., Lundrigan M., Gayda R. C., Markovitz A., Earhart C. omp T: Escherichia coli K-12 structural gene for protein a (3b). J Bacteriol. 1983 Feb;153(2):1104–1106. doi: 10.1128/jb.153.2.1104-1106.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. S., Lustig F., Borén T., Lagerkvist U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc Natl Acad Sci U S A. 1987 May;84(10):3166–3170. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susskind M. M., Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978 Jun;42(2):385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjarnardóttir S., Dingermann T., Rafnar T., Andrésson O. S., Söll D., Eggertsson G. Leucine tRNA family of Escherichia coli: nucleotide sequence of the supP(Am) suppressor gene. J Bacteriol. 1985 Jan;161(1):219–222. doi: 10.1128/jb.161.1.219-222.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman K. P., Tu C. P. Complete sequence of IS3. Nucleic Acids Res. 1985 Mar 25;13(6):2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Winans S. C., Elledge S. J., Krueger J. H., Walker G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagil E., Dolev S., Oberto J., Kislev N., Ramaiah N., Weisberg R. A. Determinants of site-specific recombination in the lambdoid coliphage HK022. An evolutionary change in specificity. J Mol Biol. 1989 Jun 20;207(4):695–717. doi: 10.1016/0022-2836(89)90238-6. [DOI] [PubMed] [Google Scholar]
- Yoshimura M., Inokuchi H., Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. IV. Amber suppressor Su+6 a double mutant of a new species of leucine tRNA. J Mol Biol. 1984 Aug 25;177(4):627–644. doi: 10.1016/0022-2836(84)90041-x. [DOI] [PubMed] [Google Scholar]