Abstract
“Natural” polyreactive antibodies, which bind in a nonspecific manner to a range of biological molecules both of self- and nonself- origin, are normal constituents of serum and are a significant part of the immune repertoire in many species, including humans. Autoantibodies to sTNF-R (the 55-kDa extracellular domain of the human receptor to tumor necrosis factor α) were affinity purified from normal human sera using immobilized sTNF-R. The isolated anti-sTNF-R IgG bound both native and denatured forms of the receptor with low affinity. These antibodies also bound to different proteins and therefore are considered to be polyreactive. We used the anti-sTNF-R antibodies and purified polyreactive antibodies to mannose-specific lectin from garlic (Allium sativum) for screening a peptide library displayed on filamentous M13 phage. After the biopanning procedure, we failed to find epitopes with a consensus sequence; however, we found that proline is the most frequent amino acid in the selected phagotopes. Proline is commonly present at solvent-exposed sites in proteins, such as loops, turns, N-terminal first turn of helix, and random coils. Thus, structures containing proline can serve as conformation-dependent common “public” epitopes for polyreactive natural antibodies. Our findings may be important for understanding polyreactivity in general and for the significance of polyreactive natural antibodies in immunological homeostasis.
Keywords: human serum, natural autoantibodies, peptide display library, polyreactivity/soluble tumor necrosis factor receptor
Natural autoantibodies, directed against a wide range of self-antigens, are present in the sera of healthy humans (1). The role of these autoantibodies is unknown. Autoimmune diseases are characterized by the presence of autoantibodies to self-proteins (2). Evidence has accumulated that indicates that self-proteins are processed and presented similarly to foreign antigens by class I and class II major histocompatibility complex (MHC) molecules (3). Self-antigenic determinants (epitopes) are presented by the class II MHC. In this context, it was shown that most (about 90%) of the class II ligands found so far originate from cellular proteins of the animal itself, mostly from plasma membrane proteins (4). Among the ligands that have been found on MHC class II are peptides derived from the transferrin receptor, Na+–K+ ATPase and from MHC class I molecules (5).
Recently, we described natural antibodies to dietary proteins in normal human serum that can be divided into two categories—specific or polyreactive (6, 7). The specific antibodies reacted with a single protein, whereas polyreactive antibodies crossreacted with a series of unrelated proteins including autoantigens (8).
One of the autoantigens that crossreacted with polyreactive natural antibodies was sTNF-R, the soluble type I (p55) truncated form of the receptor of tumor necrosis factor α (TNFα). sTNF-R exists in two different forms, type I (p55) and type II (p75), isolated from either urine or sera, from both healthy donors and cancer patients (9). These proteins function as inhibitors of TNFα activity (10).
Because sTNF-R is the cell surface portion of the membrane-bound TNF-R and circulates normally in serum, we determined whether healthy individuals have autoantibodies to sTNF-R and succeeded to purify such autoantibodies. These antibodies were shown to be polyreactive, and, using a phage display library, we determined some of the epitopes (11). For comparison, we also determined the epitopes of human natural antibodies against a lectin (ASA) from garlic, which also proved to be polyreactive (6).
We found that most of the peptides that interact with the two natural antibodies to self- and nonself-proteins contain proline. We concluded that the low affinity polyreactive antibodies are against the exposed epitopes of proteins, namely, against loops and turns. Most of the protein antigens contain proline in these sites, and this property may contribute to the polyreactive character of natural antibodies.
MATERIALS AND METHODS
Materials.
Alkaline phosphatase-conjugated goat anti-human F(ab)′, bovine serum albumin (BSA), p-nitrophenyl phosphate, biotinamidocaproate N-hydroxysuccinimide ester, and Tween 20 were from Sigma; horseradish peroxidase-conjugated goat anti-human F(ab)′ from Jackson ImmunoResearch; crosslinked Sepharose 4B and protein-A Sepharose from Pharmacia; and 96-well, flat-bottomed microtiter plates were obtained from Nunc. Avidin, conalbumin, lysozyme, ovalbumin, ovomucoid, and riboflavin-binding protein (RBP) were obtained from Belovo Chemicals (Bastogne, Belgium). Alliinase and ASA (Allium sativum agglutinin) were purified as described (6). sTNF-R was from Interpharm (Kiryat Weizmann, Israel). The hexapeptide phage-epitope library was from G. Smith (University of Missouri, Columbia) (11). Human sera were from the Israel Blood Center/Magen David Adom.
Affinity Purification of Human Anti-sTNF-R and Anti-ASA Antibodies.
Antigens were coupled to nitrophenylchloroformate-activated Sepharose (12) by incubation in PBS (pH 8) for 48 hr. Unreacted nitrophenyl groups were blocked with 100 mM NH4OH. About 2 mg of protein were bound per milliliter of resin.
Igs from 200 ml of normal human plasma, pooled from 16 healthy adult donors, were enriched by precipitation with 45% ammonium sulfate. The precipitate was dialyzed against PBS and applied to an antigen-affinity column (overnight, 4°C; recycling mode at 6 ml/min). The column was washed extensively with 50 mM Tris⋅HCl (pH 7.5), followed by the same buffer containing 0.5 M NaCl. Bound antibodies were eluted using 0.1 M glycine (pH 3), the column was brought to neutrality with PBS, and then subjected to a second elution step using 0.1 M triethylamine (pH 11). The pH of the eluted fractions was immediately neutralized with 1 M Tris⋅HCl buffer (pH 8). The antibodies, eluted either with acidic or basic buffers, were pooled and rechromatographed on a protein A column to enrich the IgG fraction.
Serology.
Antibodies were detected by ELISA. Microtiter plates were coated (100 μl per well) with antigen (10 μg/ml) in 50 mM carbonate buffer (pH 9.8) and incubated overnight at 4°C. The plates were washed three times with phosphate-buffered saline (PBS) and blocked for 2 hr at 37°C with PBS (100 μl per well) containing 3% BSA and 0.05% Tween 20 (BSA–Tween buffer). The plates were washed with PBS, and the solutions containing purified antibodies added to the antigen-coated wells (100 μl per well). Following an additional incubation for 2 hr at 37°C, the plates were washed three times with PBS, and a solution (100 μl per well) containing alkaline phosphatase-conjugated goat anti-human F(ab)′ (diluted 1:2,000) was added. After 2 hr at 37°C, the plates were washed extensively with PBS, and substrate solution (10 mg of p-nitrophenyl phosphate in 10 ml of 100 mM diethanolamine buffer, pH 9.5) was applied (100 μl per well). After 30 min, the A405 was measured using an ELISA reader.
Competition Assay.
sTNF-R was preincubated with protein A-purified human IgG in BSA–Tween buffer. After 14 hr of incubation at 4°C, the samples were applied to sTNF-R-coated wells. Bound antibodies, in the presence of different amounts of a desired competitor, were detected by ELISA as described above.
Electrophoresis and Immunoblotting.
SDS/PAGE was performed under reducing conditions using 15% acrylamide slab gels. Gels were stained with 0.05% Coomassie brilliant blue R 250 (Bio-Rad) or used for immunoblotting. For immunoblotting, sTNF-R was transferred from unstained gels to nitrocellulose sheets [150 mA for 2 hr; transfer buffer consisted of 25 mM Tris⋅HCl/192 mM glycine in methanol/water (1:5)]. The blots were blocked with BSA–Tween buffer and then incubated with affinity-purified antibodies (20 μg/ml of BSA–Tween buffer). The blots were washed three times with PBS and incubated for 2 hr with horseradish peroxidase-conjugated goat anti-human F(ab), diluted 1:10,000 in BSA–Tween buffer. After washing four times with PBS for 10 min, the blots were developed using enhanced-chemiluminescence reagent (Amersham) and exposed to x-ray film (Kodak).
Biotinylation of Anti-sTNF-R and Anti-ASA Antibodies.
For biotinylation, 200 μg of the affinity-purified human anti-sTNF-R or anti-ASA antibodies in 200 μl of 0.1 M NaHCO3 (pH 8.5) were incubated for 2 hr at room temperature with 5 μl of biotinamidocaproate N-hydroxysuccinimide ester solution in dimethylformamide (1 mg/ml). Unreacted and hydrolyzed reagent were removed by dialysis against PBS (13).
Selection of Phagotopes from the Hexapeptide Epitope Library.
A sample containing 1010 phages was mixed with 1 μg of biotinylated antibodies in 30 μl of 3% BSA/PBS solution and preincubated overnight at 4°C. The reaction mixture was then transferred into streptavidin-coated 60-mm polystyrene Petri dishes, containing 1 ml of 3% BSA/PBS, and the dishes were incubated for 30 min at room temperature. Unbound phages were removed by washing (10 times, 10 min each) in PBS/0.05% Tween 20, and the bound phages were eluted with 1 ml of glycine/HCl buffer (pH 2.2). The eluate was neutralized and used to infect Escherichia coli K91 cells. After three rounds of biopanning, individual bacterial colonies were selected and amplified for DNA sequencing (14).
DNA Sequencing.
Phages from supernatants of the selected colonies were precipitated with polyethylene glycol, and single-stranded DNA was prepared by phenol extraction. DNA sequences were determined by the dideoxy chain termination method using an Applied Biosystems (model 373A) DNA sequenator (11).
RESULTS
Antibodies to sTNF-R in Normal Human Sera.
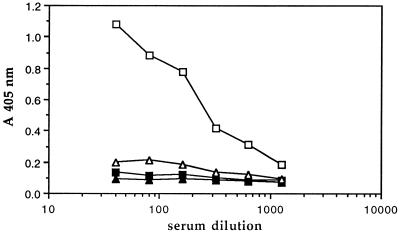
To determine whether human serum contains antibodies to the extracellular portion of sTNF-R, we applied an ELISA system in which microtiter plates were coated with the recombinant antigen (10 μg/ml), followed by total human IgG. Despite the crossreaction of sTNF-R with natural human antibodies to ASA lectins, we could not, at first, detect antibodies to sTNF-R in normal sera. Since the anti-ASA antibodies were eluted from the antigen column with acid, we considered that acid treatment of human serum may reveal antibodies to sTNF-R. Indeed, upon acidification with glycine buffer (pH 3) and subsequent neutralization, antibodies could be detected (Fig. 1). No binding could be shown using IgG-depleted serum, prepared by immunoaffinity chromatography using either an antigen-containing column or protein A-Sepharose. We therefore continued our studies with IgG purified on a protein A column.
Figure 1.
Dose-dependent binding of Igs to an sTNF-R-coated plate. Normal (▪, □) or IgG-depleted (▴, ▵) human serum was used for the binding experiment, before (▪, ▴) and after (□, ▵) preliminary acidification. Acidification was accomplished using 0.1 M glycine buffer (pH 3), and the acidified serum was immediately neutralized using 1 M Tris⋅HCl (pH 8). The level of bound antibodies was determined by ELISA.
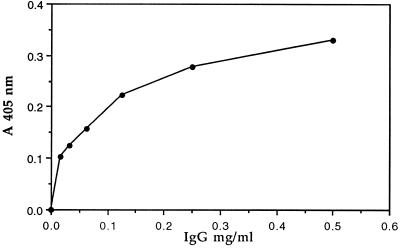
Different amounts of human IgG were introduced into sTNF-R-coated wells, and bound Igs were detected using an enzyme-labeled, anti-human antibody. Fig. 2 shows that the total human IgG contains antibodies that react with sTNF-R. Antibodies to sTNF-R were already detected at an Ig concentration of 30 μg/ml.
Figure 2.
Dependence of bound anti-sTNF-R antibodies on the total IgG concentration. The total IgG fraction was purified from normal human serum by protein A-Sepharose affinity chromatography. Solutions containing different concentrations of total human IgG in BSA–Tween buffer were added to sTNF-R-coated immunoassay plates. The level of bound Igs was monitored using enzyme-conjugated second (anti-human) antibodies.
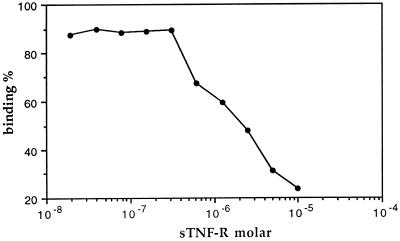
Preincubation with different amounts of sTNF-R inhibited the binding of the human IgG to the sTNF-R-coated plates. The observed inhibition was about 75% at a concentration of 10−5 M of sTNF-R (Fig. 3).
Figure 3.
Antigen-induced inhibition of anti-sTNF-R binding activity. Protein A-purified total human IgG was incubated for 14 hr at 4°C with varying concentrations of free sTNF-R, after which the samples were introduced into sTNF-R-coated wells. Detection of bound antibodies was accomplished by ELISA.
Affinity Purification of sTNF-R Antibodies.
Affinity-purified anti-sTNF-R IgG autoantibodies were obtained by applying the IgG, pooled from 16 healthy donors, to an sTNF-R-Sepharose column. The eluted antibodies were further purified on protein A-Sepharose to remove non-IgG constituents. SDS/PAGE (reducing conditions) of the eluted material revealed two major bands that corresponded to the light and heavy IgG chains (data not shown). The yield of human sTNF-R antibodies was about 120 μg from 200 ml of serum.
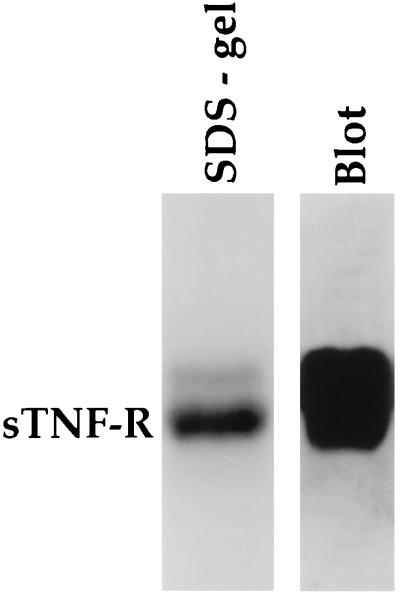
The purified antibodies bound to the TNF receptor using both ELISA and dot-blot assays. The recombinant sTNF-R was applied to SDS/PAGE and Western blotted onto nitrocellulose sheets. The affinity-purified anti-sTNF-R antibodies also recognized the denatured form of sTNF-R (Fig. 4).
Figure 4.
Identification of sTNF-R by Western blotting using affinity-purified human anti-sTNF-R antibodies. Following SDS/PAGE, the sTNF-R was transferred to nitrocellulose paper and incubated for 2 hr with affinity-purified antibodies (10 μg/ml), followed by horseradish peroxidase-conjugated second anti-human IgG.
Since the affinity-purified anti-sTNF-R antibodies interacted with both native and denatured forms of the receptor, we examined whether these antibodies might mediate any biological effect. The natural human anti-sTNF-R antibodies did not inhibit the binding of TNFα or cytotoxic monoclonal anti-sTNF-R (15) antibodies to the receptor in competitive ELISA and did not stimulate cell death in apoptotic assay (data not shown).
Polyreactivity of the Natural Autoantibodies Eluted from the sTNF-R Affinity Column.
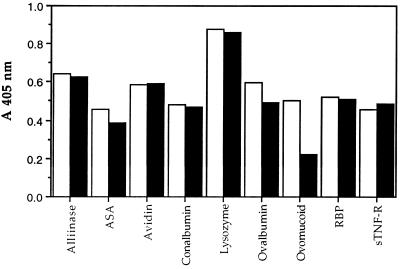
We examined the specificity of anti-sTNF-R IgG using binding experiments with many different proteins. Fig. 5 shows that the anti-sTNF-R antibodies crossreacted with all the proteins examined, similar to the reaction of the anti-ASA antibodies. Therefore, it can be concluded that these antibodies are nonspecific and polyreactive.
Figure 5.
Specificity of human antibodies eluted from ASA and sTNF-R affinity columns. Affinity-purified human anti-ASA (open columns) and human anti-sTNF-R (solid columns) antibodies (10 μg/ml) were added to wells of a microtiter plate coated with different proteins. Detection of bound antibodies was accomplished by ELISA. RBP, riboflavin-binding protein.
Selection of Bacteriophages Displaying Peptides That Bind Human Polyreactive Anti-sTNF-R and Anti-ASA Antibodies.
To determine whether certain structural elements were preferable for binding by polyreactive antibodies, we performed epitope mapping experiments using a random hexapeptide phage library (11).
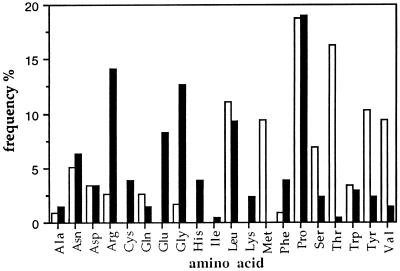
Table 1 shows the sequences of displayed peptides selected after the third round of biopanning on anti-sTNF-R and anti-ASA antibodies. We found that Pro is the most frequent residue in the selected phagotopes, whereas Ala, Ile, Gln, and Lys are the least frequent residues (Fig. 6).
Table 1.
Sequences of peptides selected using the polyreactive human anti-sTNF-R and anti-ASA antibodies
| Peptides selected using human anti-sTNF-R antibodies | Peptides selected using human anti-ASA antibodies | ||
|---|---|---|---|
| RRFFLV | 1 | PLYTVM | 11 |
| SRRFLG | 1 | WPDNPT | 4 |
| LREFPH | 1 | PTGSNS | 2 |
| RREFPG | 4 | PRYFQA | 1 |
| DPGSG | 3 | RTQLSS | 2 |
| KPPGYG | 2 | ||
| NCCGEG | 4 | ||
| QENPDK | 3 | ||
| WELPRN | 2 | ||
| PHWHWE | 1 | ||
| PPWHLP | 4 | ||
| RRYLRP | 2 | ||
| RIYLRP | 1 | ||
| NRLPHL | 2 | ||
| NPPELA | 2 | ||
| RALVRG | 1 | ||
| RGVQSR | 1 | ||
| TDSKKK | 1 | ||
| Total | 36 | 20 | |
The values represent the number of phages sequences. The corresponding phage epitopes (single-letter amino acid code) were deduced from the DNA sequence.
Figure 6.
The frequency of amino acids in the peptides selected from the phage-display library. The frequency was calculated using the sequences deduced from the 36 phagotopes selected with the human anti-sTNF-R antibodies (solid columns) and from the 20 phagotopes selected with the human anti-ASA antibodies (open columns).
We used results obtained from protein crystallography as described in ref. 16 to calculate the probability of occurrence of -X-P- pairs (where X is any amino acid) in different structural elements of the proteins. Table 2 summarizes the results of our estimation, which support the exposed location of these proline-containing sequences in the proteins studied.
Table 2.
The probability of the existence of aa-Pro pairs from selected peptides within secondary structural elements
| aa-Pro | aa-Pro pair, n | α-Helix | β-Sheet | Turn | Loops, random coils |
|---|---|---|---|---|---|
| Ala-Pro | 15 | 24.4 | 15.4 | 19.2 | 41.0 |
| Arg-Pro | 3 | 13.9 | 19.4 | 30.6 | 36.1 |
| Asn-Pro | 7 | 42.9 | — | 23.2 | 33.9 |
| Asp-Pro | 3 | 50.0 | 0.9 | 30.3 | 18.8 |
| Leu-Pro | 8 | 21.8 | 12.6 | 25.3 | 40.3 |
| Phe-Pro | 5 | 25.6 | 14.0 | 18.6 | 41.8 |
| Pro-Pro | 8 | 21.6 | 5.4 | 24.3 | 48.7 |
| Trp-Pro | 4 | 33.3 | 11.1 | 22.2 | 33.4 |
| Average probability of location | 27.8 | 10.1 | 23 | 39.1 |
DISCUSSION
We describe the isolation and properties of purified human natural autoantibodies from healthy individuals to the receptor for TNFα. Antibodies against the soluble form of this receptor (sTNF-R) were purified from normal human serum. Anti-sTNF-R antibodies recognize both the native and denatured form of the receptor. The antibodies were polyreactive and of low affinity. These properties make them similar to other human natural antibodies against self-antigens (17, 18) as well as to several dietary proteins, such as the mannose-specific lectin from garlic (6).
Only low numbers of antibodies to the sTNF-R receptor could be detected in normal human serum. However, the amount of antibody activity increased significantly (5- to 6-fold) when the serum was acidified. This phenomenon has also been observed with other autoantigens (6, 19). This may indicate that, in normal human serum, antibodies can associate with one another, perhaps by an anti-idiotypic cascade or by combining with other antigens that prevent them from interacting with self-antigens under normal conditions. When the serum is passed through a protein A immunoaffinity column, the effluent fractions fail to interact with sTNF-R, indicating that the observed binding is mediated via IgGs and not by TNF or other proteins present in the serum. Interestingly, natural autoantibodies from healthy individuals, isolated on acetylcholine- or γ-interferon-receptor columns, behave similarly and are also polyreactive (unpublished results). The natural antibodies did not interfere with the regular functions of sTNF-R.
The finding that many human natural antibodies are polyreactive and crossreact with self- and nonself-antigens has led to many theories regarding the molecular basis of such crossreactivity. Kabat et al. (18) suggested the involvement of spatial distribution of charges, while others suggested that that autoantibodies recognize common structures present on phylogenetically conserved proteins (20). We therefore decided to use a hexapeptide phage-display library (11) to determine whether there are common epitopes recognized by two different types (i.e., anti-sTNF-R and anti-ASA) of natural polyreactive human antibodies.
Epitope mapping experiments suggested a general lack of specificity in this system, because analysis of the sequences of 56 random phages that bound to both antibodies failed to reveal a consensus sequence. Analysis of the frequency of the relative occurrence of the amino acids in these phages indicated that proline was the most frequently occurring amino acid in these peptides (Fig. 6). Interestingly, epitope mapping of naturally occurring autoantibodies against TNF-α also revealed the prominence of proline and serine (21). These findings clearly indicate that human natural polyreactive antibodies may not recognize linear epitopes but may recognize conformation-dependent epitopes involving the sequence -X-Pro-X- (where X is any amino acid).
Proline-containing peptides have indeed been connected to many immunologically important phenomena. Autoantibodies from many patients with systemic lupus erythematosus bind the Sm autoantigen B/B′ polypeptide. In this system, proline-containing peptides are early targets of the autoimmune response (22). In another disease, the autoantigen of type-II mixed cryoglobulinemia is a peptide with a consensus sequence that contains proline. The same sequence is present in the LAG-3 protein on the surface of lymphocytes and is suggested to be an exposed epitope (23, 24). In addition, the neutralizing determinant of human immunodeficiency virus type I, which elicits neutralizing antibodies, is an eight amino acid peptide of the V3 loop containing a central conserved sequence Gly-Pro-Gly (25–27). Finally, Tax peptides, which were used to crystallize the T cell antigen receptor with the MHC, also contain proline (28).
Many of the self-peptides that are similar to viral T cell epitopes that cause autoimmunity contain proline at different positions (29). The two peptides of the human acetylcholine receptor α-subunit, which bind HLA class II of patients with myasthenia gravis, contain proline. In contrast, a peptide from the same region of the same subunit did not contain proline and failed to bind (30). Autoantibodies against the nonhistone nucleosomal protein, HMG-17, were shown to recognize an eight residue peptide containing proline and cysteine (31). Most of the epitopes on proteinase-3, recognized by antibodies from patients with Wegner’s granulomatosis, contain proline (32).
There is a long list of known proline-containing epitopes on autoantigens. This is not surprising since proline commonly occurs in proximity to protein–protein interaction sites (33). Proline is also important for chain conformation and protein folding, and the sequences containing proline tend to be conserved. The unique nature of proline and its tendency to appear in solvent-exposed regions of proteins probably contribute to their reputed immunogenic properties.
Proline-rich motifs are also present in pathogenic bacteria and viruses (34), and therefore polyreactive low-affinity antibodies against proline-rich sequences may contribute to the first line of defense against foreign invaders. Since proline is present in most proteins at loops, turns, random coils, and in the initial turn of a helix, the immune system may recognize these structures as self-antigens and “mark” them for further processing.
The physiological role of autoantibodies in healthy people and autoimmune patients is not known. One possibility is that these antibodies may induce autoimmunity upon pathological deviation from normal physiological function (35) as suggested originally by Grabar (36) in accordance with recent studies of Avrameas and Ternynck (37).
Acknowledgments
We thank Prof. D. Wallach for fruitful discussions and Dr. M. Boldin and A. Dibman for help during our study. We also thank Dr. E. A. Bayer for critical reading of the manuscript. This work was supported by research grants from the Lynne and William Frankel Fund for the Diagnosis and Treatment of Ovarian and Breast Cancer and MINERVA, Deutsch-Israelische Wissenschftliche Zusammenarbeit, Germany.
ABBREVIATIONS
- ASA
Allium sativum agglutinin
- TNFα
tumor necrosis factor α
- sTNF-R
55-kDa extracellular domain of the human receptor for TNFα
- MHC
major histocompatibility complex
References
- 1.Avrameas S, Guilbert B, Dighiero G. Ann Immunol. 1981;132:231–236. doi: 10.1016/0769-2625(81)90031-3. [DOI] [PubMed] [Google Scholar]
- 2.Schattner A. Immunol Lett. 1987;14:143–153. doi: 10.1016/0165-2478(87)90094-0. [DOI] [PubMed] [Google Scholar]
- 3.Moudgil K D, Ametani A, Grewal I S, Kumar V, Sercarz E E. Int Rev Immunol. 1993;10:365–377. doi: 10.3109/08830189309061711. [DOI] [PubMed] [Google Scholar]
- 4.Rotzschke O, Falk K. Curr Opin Immunol. 1994;6:45–51. doi: 10.1016/0952-7915(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 5.Chicz R M, Urban R G, Gorga J C, Vignali D A A, Lane W S, Strominger J L. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tchernychev B, Rabinkov A, Mirelman D, Wilchek M. Immunol Lett. 1995;47:53–57. doi: 10.1016/0165-2478(95)00067-f. [DOI] [PubMed] [Google Scholar]
- 7.Tchernychev B, Wilchek M. FEBS Lett. 1996;397:139–142. doi: 10.1016/s0014-5793(96)01154-4. [DOI] [PubMed] [Google Scholar]
- 8.Sigounas G, Kolaitis N, Monell-Torrens E, Notkins A L. J Clin Immunol. 1994;14:375–381. doi: 10.1007/BF01546322. [DOI] [PubMed] [Google Scholar]
- 9.Aderka D, Engelmann H, Hornik V, Skornic Y, Levo Y, Wallach D, Kushtai G. Cancer Res. 1991;51:5602–5607. [PubMed] [Google Scholar]
- 10.Engelmann H, Aderka D, Rubinstein M, Rotman D, Wallach D. J Biol Chem. 1989;264:11974–11980. [PubMed] [Google Scholar]
- 11.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 12.Wilchek M, Miron T. Biochem Int. 1982;4:629–635. [Google Scholar]
- 13.Bayer E A, Wilchek M. Methods Enzymol. 1990;184:138–160. doi: 10.1016/0076-6879(90)84268-l. [DOI] [PubMed] [Google Scholar]
- 14.Yayon A, Aviezer D, Safran M, Gross J L, Heldman Y, Cabilly S, Givol D, Katchalski-Katzir E. Proc Natl Acad Sci USA. 1993;90:10643–10647. doi: 10.1073/pnas.90.22.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelmann H, Holtmann H, Brakebusch C, Avni Y S, Sarov I, Nophar Y, Hadas E, Leitner O, Wallach D. J Biol Chem. 1990;265:14497–14504. [PubMed] [Google Scholar]
- 16.MacArthur M W, Thornton J M. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 17.Hurez V, Dietrich G, Kaveri S-V, Kazatchkine M D. Scand J Immunol. 1993;38:190–196. doi: 10.1111/j.1365-3083.1993.tb01712.x. [DOI] [PubMed] [Google Scholar]
- 18.Kabat E A, Nickerson K G, Liao J, Grossbard L, Osserman E F, Glickman E, Chess L, Robbins J B, Schneerson R, Yang Y. J Exp Med. 1986;164:642–654. doi: 10.1084/jem.164.2.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurez V, Kaveri S-V, Kazatchkine M. Eur J Immunol. 1993;23:783–789. doi: 10.1002/eji.1830230402. [DOI] [PubMed] [Google Scholar]
- 20.Avrameas S. Immunol Today. 1991;12:154–159. doi: 10.1016/S0167-5699(05)80045-3. [DOI] [PubMed] [Google Scholar]
- 21.Sioud M, Dybwad A, Jespersen L, Suleyman S, Natvig J B, Førre ⊘. Clin Exp Immunol. 1994;98:520–525. doi: 10.1111/j.1365-2249.1994.tb05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James J A, Gross T, Scofield R H, Harley J B. J Exp Med. 1995;181:453–461. doi: 10.1084/jem.181.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortese R, Monaci P, Luzzago A, Santini C, Bartoli F, Corteze I, Fortugno P, Galfre G, Nicosia A, Felici F. Curr Opin Biotechnol. 1996;7:616–621. doi: 10.1016/s0958-1669(96)80072-3. [DOI] [PubMed] [Google Scholar]
- 24.Mecchia M, Cassato M, Filocamo G, Bonomo L, Fiorilli M, Cortese R, Migliaccio G, Nicosia A. J Immunol. 1996;157:3727–3736. [PubMed] [Google Scholar]
- 25.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 26.Trkola A, Dragic T, Arthos J, Benley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 27.Javaherian K, Langlois A J, McDanal C, Ross K L, Eckler L I, Jellis C L, Profy A T, Rusche J R, Bolognesi D P, Putney S D, Matthews T J. Proc Natl Acad Sci USA. 1989;86:6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 29.Wucherpfennig K W, Strominger J L. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zisman E, Brautbar C, Sela M, Abramsky O, Battat S, Kirshner S L, Katz-Levy Y, Dayan M, Mozes E. Hum Immunol. 1995;44:121–130. doi: 10.1016/0198-8859(95)00094-1. [DOI] [PubMed] [Google Scholar]
- 31.Neur G, Bautz F A, Bustin M, Michels H, Truckenbrodt H. Autoimmunity. 1994;17:23–30. doi: 10.3109/08916939409014655. [DOI] [PubMed] [Google Scholar]
- 32.Williams R C, Jr, Staud R, Malone C C, Payabyab J, Byres L, Underwood D. J Immunol. 1994;152:4722–4737. [PubMed] [Google Scholar]
- 33.Kini R M, Evans H J. Biochem Biophys Res Commun. 1995;212:1115–1124. doi: 10.1006/bbrc.1995.2084. [DOI] [PubMed] [Google Scholar]
- 34.Bliska J. Chem Biol. 1996;3:7–11. doi: 10.1016/s1074-5521(96)90076-9. [DOI] [PubMed] [Google Scholar]
- 35.Del Guercio P. Immunol Res. 1993;12:168–182. doi: 10.1007/BF02918302. [DOI] [PubMed] [Google Scholar]
- 36.Grabar P. Immunol Today. 1983;4:337–340. doi: 10.1016/0167-5699(83)90169-X. [DOI] [PubMed] [Google Scholar]
- 37.Avrameas S, Ternynck T. Res Immunol. 1995;146:235–248. doi: 10.1016/0923-2494(96)80259-8. [DOI] [PubMed] [Google Scholar]