Abstract
Recent identification of a C3-like gene in sea urchins revealed the presence of a complement system in invertebrates. To elucidate further the components and function of the pre-vertebrate complement system, we attempted to isolate an ascidian (urochordata) C3 convertase. After identification of C3 cDNA from Halocynthia roretzi, a Japanese ascidian, reverse transcriptase–PCR amplification of hepatopancreas RNA was performed using primers encoding highly conserved amino acid sequences of the vertebrate Bf and C2 serine protease domain. Two candidate sequences were identified, and the corresponding cDNA clones were isolated from a hepatopancreas library. Surprisingly, neither clone is related to Bf/C2 but rather share the same domain structure of mammalian C1r/C1s/MASP (mannan binding protein-associated serine protease), and are more related evolutionarily to mammalian MASP than to mammalian C1r or C1s. The identification of the tunicate MASP clones, amplified with primers designed to amplify Bf or C2, suggests that the lectin pathway antedated the classical and alternative pathways of complement activation.
Keywords: evolution, tunicate
The mammalian complement system plays a pivotal role in host defense against infection and is organized in two activation pathways, designated classical and alternative, and in the lytic pathway of membrane attack (1). The key event in the system is proteolytic activation of the central component C3 into C3b, which covalently tags microorganisms to be cleared from circulation or tissues. The C3 convertases of the classical and alternative pathways are composed of C2 and C4, Bf and C3, respectively. Gene duplications are believed to be responsible for generation of these two activation pathways, because C2 and Bf as well as C4 and C3 are homologous proteins.
Evolutionary studies have revealed the presence of the complement system in all major classes of vertebrates (2). The complement system of the lamprey, a representative of the most primitive class of extant vertebrates, appears to consist only of the alternative pathway, and functions as an opsonic system (3). In contrast, all gnathostomes have well developed complement systems with both the alternative and classical activation pathways and the lytic pathway (2). Recently, a cDNA clone with significant sequence identity to vertebrate C3, C4, and C5 has been identified in the sea urchin, a marine invertebrate (4), placing the phylogenetic origin of the complement system in the invertebrate line.
The recent finding that mannan (or mannose) binding protein (MBP) and MBP-associated serine protease (MASP, also called P100) can activate the complement system in an antibody-independent fashion (5–7) revealed the existence of a third activation mechanism of the mammalian complement system, designated the lectin pathway (8). In this pathway, the C-type lectin domain of MBP recognizes mannose or N-acetylglucosamine structures on the surfaces of pathogens, leading to the activation of MASP, which is present as a proenzyme complexed with MBP. The activated MASP then activates the classical pathway by proteolytically cleaving C4 and C2. MBP and MASP show structural similarity to C1q and C1r/C1s, respectively, indicating that gene duplication events were also responsible for generating the classical and lectin pathways. Although the details of the mammalian lectin pathway are not well known, demonstration of a lectin-based recognition system suggests an ancient origin for this pathway. Supporting this idea, MBP has been isolated from chicken serum (9) and MASP cDNA clones have been identified from Xenopus, carp, and lamprey (Yuichi Endo, personal communication). However, neither MBP nor MASP has been identified in invertebrates.
Tunicates occupy a pivotal intermediary position between invertebrates and vertebrates. After isolation of C3 cDNA from the Japanese ascidian, Halocynthia roretzi (unpublished observation), we attempted to isolate cDNA encoding the C3-activating enzyme, Bf, or C2. Characterization of candidate clones, however, indicated that they were not Bf or C2 but rather MASP. Identification of MASP in tunicates suggests that the lectin pathway was the original mechanism for activation of the complement system.
MATERIALS AND METHODS
Materials.
Solitary ascidians, Halocynthia roretzi, were harvested in Mutsu Bay, Japan. The hepatopancreas was removed from dissected ascidians immediately before use. Hemocytes were obtained as described previously (10) by centrifugation from hemolymph collected by cutting the tunic matrix without injuring the internal organs. Restriction enzymes were purchased from Toyobo (Osaka) and New England Biolabs. The ligation kit was from Takara Shuzo (Kyoto). [α-32P]dCTP, the Rediprime Random Primer Labeling kit, and cDNA Synthesis Systems Plus were from Amersham Japan (Tokyo). λ Zap II and Gigapack Gold were from Stratagene. DNA Sequencing System 373A Analysis Software version 1.01 and the Prism Dye Terminator Cycle Sequencing kit were from Applied Biosystems Japan. The EcoRI adapter was from Promega. The TA cloning kit was from Invitrogen.
RNA Extraction and cDNA Library Construction.
RNA was isolated from hepatopancreas and hemocytes using guanidine thiocyanate (11), and poly(A)+ RNA was selected on an oligo(dT)-cellulose column (12). Construction of an ascidian hepatopancreas cDNA library was carried out as described previously (13), and approximately 3 × 105 plaques were screened.
Reverse Transcriptase–PCR (RT-PCR) Amplification of Candidate mRNA Segments for Ascidian Bf/C2.
Degenerate PCR primers were the same as those used for the amplification of lamprey (14) and Xenopus Bf (13) (Fig. 1). PCR template was double-stranded cDNA synthesized from ascidian hepatopancreas and hemocyte mRNA using cDNA Synthesis Systems Plus. Thirty cycles of amplification were carried out in an Astec PC700 thermocycler (Fukuoka, Japan) using the following parameters: 94°C for 0.5 min, 50°C for 1 min, and 72°C for 1 min (15). PCR products of the expected size (250 bp) were gel-purified and ligated into the pCRII vector (Invitrogen).
Figure 1.
RT-PCR amplification of the Bf/C2-like serine protease domain from Halocynthia roretzi. RT-PCR primers were prepared using the amino acid sequences in the serine protease domain, which are highly conserved among all Bf and C2 analyzed to date. Two different clones were isolated and designated Hr1 and Hr2. The deduced amino acid sequences of these clones were aligned with the corresponding regions of human Bf (HuBf) and C2 (HuC2). The three diagnostic residues that are completely conserved among all Bf and C2 are indicated by ∗. The first residue of Hr1 is shown here as F; however, the amino acid at this position deduced from a full-length cDNA clone was V. This discrepancy was due to the fact that the first two nucleotides encoding this amino acid of the RT-PCR product had originated from the 5′ primer, and the template sequence contained a mismatch at the first nucleotide (see text).
Nucleotide Sequence Analysis.
DNA sequence analysis was performed by the dideoxy chain termination method (16) using an Applied Biosystems 373A DNA sequencer. Sequencing primers were synthesized to extend sequence readings using an Applied Biosystems 381A DNA synthesizer. Each sequence was determined at least twice from both strands.
Northern Blotting Analysis.
Total RNA from ascidian hepatopancreas and hemocytes was denatured by glyoxal, separated on a 1% agarose gel, and blotted onto a nylon membrane (Hybond-N, Amersham) (17). Hybridization with radiolabeled probes prepared using the Rediprime kit (Amersham) was performed in 10× Denhardt’s solution/1 M sodium chloride/50 mM Tris/10 mM EDTA/0.1% SDS/0.1 mg/ml denatured salmon sperm DNA at 65°C for 16–20 hr. Membranes were washed twice for 30 min at 65°C in 0.1× standard saline citrate and 0.1% SDS.
RESULTS AND DISCUSSION
RT-PCR Amplification of Bf/C2-like mRNA.
RT-PCR amplification of the ascidian hepatopancreas and hemocyte mRNA using primers corresponding to the two highly conserved amino acid sequences in the serine protease domain of Bf and C2 resulted in a single DNA band of the expected size (about 250 bp). The DNA was gel-purified and subcloned into the plasmid using the TA cloning kit. Twenty hepatopancreas clones and 16 hemocyte clones were randomly selected, and nucleotide sequences of the inserts were determined. Candidates for Bf/C2 were initially screened for three amino acid residues that are completely conserved in all Bf/C2 sequences analyzed to date (indicated by ∗ in Fig. 1). Two different Bf/C2-like sequences, referred to as Hr1 and Hr2, were identified among other serine protease sequences. Hr1 possessed all three diagnostic residues and was represented in one hepatopancreas clone and two hemocyte clones. Hr2 possessed two of the diagnostic residues and was represented in six hepatopancreas clones. Amino acid identities of Hr1 and Hr2 to the corresponding region of human Bf were 16% and 14%, respectively, and identity between Hr1 and Hr2 was 13%.
Identification of the Two Ascidian Sequences as MASP.
Using Hr1 and Hr2 as probes, the ascidian hepatopancreas cDNA library containing 3 × 105 independent clones was screened, and eight Hr1- and 14 Hr2-hybridizing clones were isolated. Clones from each group were shown to have the same restriction enzyme maps. The clones with the longest insert in each group, 2,572 bp for Hr1 and 2,526 bp for Hr2, were sequenced in their entirety. As will be discussed below, these clones seemed to encode common ancestral molecules of mammalian MASP and C1r/C1s, which presumably showed a closer similarity to mammalian MASP than to mammalian C1r/C1s, and were designated AsMASPa and AsMASPb. AsMASPa contained a 2,572-bp-long insert including a 17-bp poly(A)+ tail (Fig. 2). A 182-bp 5′ untranslated region is followed by a 2,241-bp ORF, including a stop codon and a 132-bp 3′ untranslated region. AsMASPb contained a 2,526-bp-long insert with a 101-bp 5′ untranslated region followed by a 2,259-bp ORF, including a stop codon and a 147-bp 3′ untranslated region.
Figure 2.
Nucleotide and deduced amino acid sequences of AsMASPa and AsMASPb. The complete nucleotide and deduced amino acid sequences of AsMASPa (A) and AsMASPb (B) are shown. Nucleotide and amino acid residue numbers of the rightmost residues of each line starting from the initiation methionine codon are shown at the right. The possible poly(A)+ addition signals are underlined. Stars indicate the stop codons.
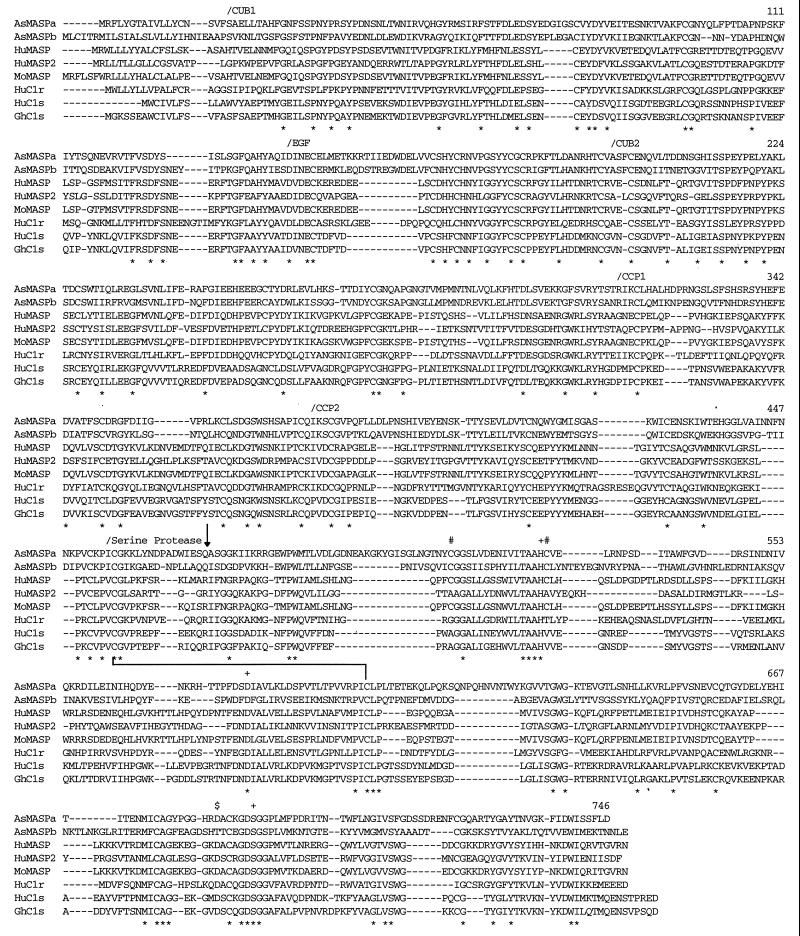
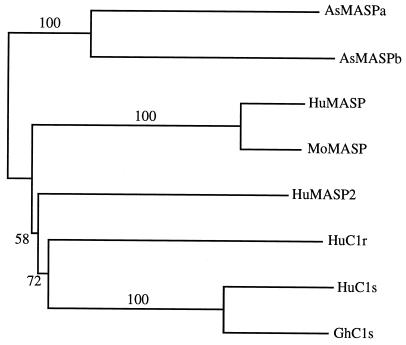
A homology search of the deduced amino acid sequences of AsMASPa and AsMASPb indicated that these sequences show a close structural similarity to mammalian C1r/C1s/MASP. Therefore, a multiple alignment of these sequences with human C1r (18), human C1s (19), golden hamster C1s (20), human MASP (8), mouse MASP (21), and recently identified human MASP2 (22) was performed using Clustal W software (23). The AsMASPa and AsMASPb amino acid sequences show a significant similarity to the mammalian C1r, C1s, and MASP sequences throughout their entire length (Fig. 3). Thus, the domain structure characteristic of mammalian C1r/C1s/MASP, which stretches from the N terminus, CUB, EGF, CUB, CCP, CCP, and serine protease domains (8, 21, 24), is found in the two ascidian sequences. The calculated amino acid identities for AsMASPa based on the above alignment are 44% with AsMASPb, 27% with human MASP, 28% with mouse MASP, 28% with human MASP2, 27% with human C1r, 26% with human C1s, and 28% with hamster C1s. The identities of AsMASPb are 28% with human MASP, 28% with mouse MASP, 28% with human MASP2, 26% with human C1r, 24% with human C1s, and 24% with hamster C1s. Thus, it is difficult to assign a MASP or C1r/C1s idenitity to AsMASPa and AsMASPb by simply comparing overall amino acid identities. Instead, nearly the same genetic distance of the ascidian sequences from mammalian MASP and C1r/C1s strongly suggests that AsMASPa and AsMASPb represent a common ancestral state of mammalian MASP and C1r/C1s. The higher amino acid identities between mammalian MASP and C1r/C1s (34% to 37%) indicated that the gene duplication event that generated MASP and C1r/C1s from a common ancestor occurred in the vertebrate lineage. However, AsMASPa and AsMASPb have certain structural features characteristic of mammalian MASP: the two cysteine residues (512C and 528C)§ involved in histidine loop formation of mammalian MASP but which are absent from human MASP2 (22) and mammalian C1r or C1s (25) are present in both the ascidian sequences. Moreover, the codons encoding the active site Ser residue (690S) of the two ascidian sequences are TCG and TCT, respectively (Fig. 2), and the corresponding codons of mammalian MASP and C1r/C1s/human MASP2 are TCN and AGY, respectively (8, 22, 26). We therefore designate the two ascidian sequences AsMASPa and AsMASPb, and suggest that the common ancestor of MASP and C1r/C1s/MASP2 was more similar to MASP than to C1r/C1s/MASP2. A phylogenetic tree constructed using the neighbor-joining method (27) based on alignment shown in Fig. 3 indicated that AsMASPa and AsMASPb form a cluster, suggesting that duplication between these two genes is a recent event that most probably occurred in the tunicate lineage (Fig. 4). Although this tree is unrooted, the branching pattern supports the hypothesis described above that gene duplication of MASP and C1r/C1s/MASP2 occurred in the vertebrate lineage. Structurally, human MASP2 is more closely related to C1r or C1s than to MASP, and there is no orthology between two ascidian MASPs and human MASP and MASP2.
Figure 3.
Alignment of AsMASPa and AsMASPb amino acid sequences with mammalian MASP, C1r, and C1s. Multiple alignment of the amino acid sequences was performed using Clustal W software. Gaps introduced to increase identity are shown by dashes. The completely conserved residues among these eight sequences are indicated by ∗ below the sequences. The amino acid numbers of the rightmost residues of AsMASPa are shown for each lane. The N-termini of each domain are marked by/above the sequences. Three residues at the catalytic site of serine proteases 527H, 579D, and 690S, two cysteine residues involved in histidine loop formation (512C and 528C), and the residue at the S1 specificity crevice (684D) are marked by +, #, and $, respectively. The arrow pointing downward indicates a processing site of mammalian MASP, C1r, and C1s. The line connecting 456C and 599C indicates a disulfide bond between two subunit chains.
Figure 4.
Phylogenetic tree of MASP, C1r, and C1s. The relationships among two MASPs of Halocynthia roretzi, human MASP (HuMASP), human MASP2 (HuMASP2), mouse MASP (MoMASP), human C1r (HuC1r), human C1s (HuC1s), and golden hamster C1s (GhC1s) were analyzed by the neighbor-joining method along their entire amino acid sequences based on alignment shown in Fig. 3. Numbers on branches are bootstrap percentages supporting a given partitioning.
When compared with mammalian MASP, human MASP2, and C1r/C1s, both AsMASPa and AsMASPb have a high degree of conservation of residues considered to be important for MASP function, such as the serine protease active site residues, 528H, 579D, and 690S, and the cysteine residues involved in disulfide bonding between the subunit chains generated by proteolytic processing (456C and 599C). However, curious replacements are observed at the putative processing site of both AsMASPa and AsMASPb: the conserved RIF sequence of mammalian MASP is replaced by 471QAS in AsMASPa and QIS in AsMASPb. These sites would not be cleaved, either autocatalytically or mutually, by enzymes with trypsin-like substrate specificity. AsMASPa, however, has a 476KII sequence five residues C-terminal to the QAS sequence, which could serve as a processing site. Moreover, it is interesting to note that the 684D residue at the S1 specificity crevice (28) of trypsin-like serine proteases is replaced by T in AsMASPb. This substitution may change the substrate specificity of AsMASPb, and there is a possibility that AsMASPb can undergo autocatalytic activation at the QIS site. In this context, it is interesting to note that the LXR sequence in the C3 convertase cleavage site, present in all C3 sequences determined to date, is not found at the corresponding position in Halocynthia roretzi C3 (unpublished observation). Thus, it is unlikely that Halocynthia roretzi C3 is activated by an enzyme similar to vertebrate C3 convertases. Therefore, both AsMASPa (with trypsin-like specificity) and AsMASPb (which lacks this specificity) might proteolytically activate C3. Biochemical analyses are required to clarify the physiological substrates and activation mechanisms of AsMASPa and AsMASPb.
Northern Blotting Analysis.
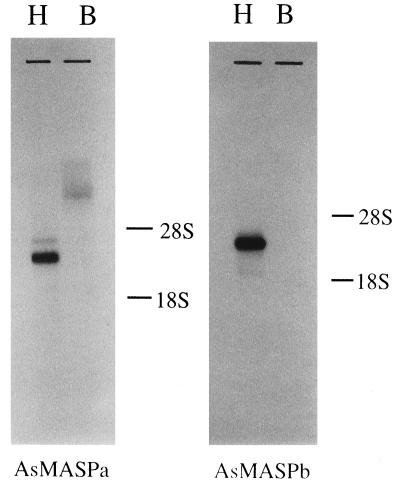
Northern blotting analysis of AsMASPa and AsMASPb was performed using two possible sources of plasma proteins, blood cells and hepatopancreas (Fig. 5). Both AsMASPa and AsMASPb probes detected a major hybridizing band from hepatopancreas. The sizes of AsMASPa and AsMASPb messages were 3.2 kb and 3.6 kb, respectively, indicating that their 5′ untranslated regions (approximately 0.6 to 1.0 kb) are still to be isolated.
Figure 5.
Northern blotting analysis of AsMASPa and AsMASPb. Five micrograms of hepatopancreas (H) and blood cell (B) RNA were denatured by glyoxal, separated on an agarose gel, and transferred to a nylon membrane. The hybridization probes used were PstI (nucleotide position 495 of Fig. 2A) to the 3′ end fragment of AsMASPa, and EcoRV (nucleotide position 816 of Fig. 2B) to the 3′ end fragment of AsMASPb.
Evolutionary Implications.
The original PCR primers used in this study were designed to amplify Bf/C2; amplification of Xenopus and lamprey liver mRNA with these primers resulted in preferential amplification of Bf (13, 14). MASP clones were never detected in spite of the presence of both Bf and MASP mRNA in these livers (Y. Endo, personal communication). Reexamination of the AsMASPa sequence indicates the presence of two mismatches each with both the sense and antisense primer sequences. Similarly, the AsMASPb sequence has mismaches with both the primers (1 sense and 4 antisense). These results strongly argue against, although do not exclude completely, the presence of Bf or C2 in tunicates. If Bf and C2 are actually absent in tunicates, the lectin pathway was likely to be the original activation pathway of the complement system. Because mammalian MASP is reported to activate C3 directly (29, 30), although the efficiency is not very high, it is conceivable that the primitive complement system consisted of MBP, MASP, and C3 and functioned in an opsonic manner. The major developments of the complement system, therefore, seem to have occurred in two stages; first, with the emergence in cyclostomes of the alternative pathway to establish a means of amplification, and second, after the divergence of cyclostomes but before the divergence of cartilaginous fish, the emergence of the classical and lytic pathways. This hypothesis would not only explain the evolution of the complement system, but also may help to reevaluate the activation mechanism of the mammalian alternative pathway. The lectin and alternative pathways may be two components of a single process of activation, in which the former plays a role as the initiator and the latter plays a role as an amplifier in the innate immune system.
Acknowledgments
We thank Ms. Chisato Yamada for technical assistance, Drs. Martin F. Flajnik, Misao Matsusita, and Teizo Fujita for helpful discussions, Drs. Jens C. Jensenius and Yuichi Endo for providing unpublished data, and Dr. William Campbell for English editing. This work was partially supported by a Grant in Aid for Scientific Research on Priority Areas (Intracellular Proteolysis and Molecular Evolution).
ABBREVIATIONS
- MBP
mannan binding protein
- MASP
MBP-associated serine protease
- RT-PCR
reverse transcriptase–PCR
Footnotes
Positions of amino acid residues are referred by the residue numbers of AsMASPa throughout this report.
References
- 1.Müller-Eberhard H J. Annu Rev Biochem. 1988;57:321–347. doi: 10.1146/annurev.bi.57.070188.001541. [DOI] [PubMed] [Google Scholar]
- 2.Dodds A W, Day A J. In: Complement in Health and Disease. Whaley K, Loos M, Weiler J M, editors. Boston: Kluwer; 1993. pp. 39–88. [Google Scholar]
- 3.Nonaka M, Fujii T, Kaidoh T, Natsuume-Sakai S, Nonaka M, Yamaguchi N, Takahashi M. J Immunol. 1984;133:3242–3249. [PubMed] [Google Scholar]
- 4.Smith L C, Chang L, Britten R J, Davidson E H. J Immunol. 1996;156:593–602. [PubMed] [Google Scholar]
- 5.Ikeda K, Sannoh T, Kawasaki N, Kawasaki T, Yamashina I. J Biol Chem. 1987;262:7451–7454. [PubMed] [Google Scholar]
- 6.Matsushita M, Fujita T. J Exp Med. 1992;176:1497–1502. doi: 10.1084/jem.176.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji Y-H, Fujita T, Hatsuse H, Takahashi A, Matsushita M, Kawakami M. J Immunol. 1993;150:571–578. [PubMed] [Google Scholar]
- 8.Sato T, Endo Y, Matsushita M, Fujita T. Int Immunol. 1994;6:665–669. doi: 10.1093/intimm/6.4.665. [DOI] [PubMed] [Google Scholar]
- 9.Laursen S B, Hedemand J E, Tiel S, Willis A C, Skriver E, Madsen P S, Jensenius J C. Glycobiology. 1995;5:553–561. doi: 10.1093/glycob/5.6.553. [DOI] [PubMed] [Google Scholar]
- 10.Azumi K, Satoh N, Yokosawa H. J Exp Zool. 1993;265:309–316. [Google Scholar]
- 11.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 12.Aviv H, Leder P. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato Y, Salter-Cid L, Flajnik M F, Kasahara M, Namikawa C, Sasaki M, Nonaka M. J Immunol. 1994;153:4546–4554. [PubMed] [Google Scholar]
- 14.Nonaka M, Takahashi M, Sasaki M. J Immunol. 1994;152:2263–2269. [PubMed] [Google Scholar]
- 15.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 16.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas P S. Proc Natl Acad Sci USA. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Journet A, Tosi M. Biochem J. 1986;240:783–787. doi: 10.1042/bj2400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tosi M, Duponchel C, Meo T, Julier C. Biochemistry. 1987;26:8516–8524. doi: 10.1021/bi00400a004. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita H, Sakiyama H, Tokunaga K, Imajoh-Ohmi S, Yamada Y, Isono K, Sakiyama S. FEBS Lett. 1989;250:411–415. doi: 10.1016/0014-5793(89)80766-5. [DOI] [PubMed] [Google Scholar]
- 21.Takayama Y, Takada F, Takahashi A, Kawakami M. J Immunol. 1994;152:2308–2316. [PubMed] [Google Scholar]
- 22.Thiel S, Vorup-Jensen T, Stover C M, Schwaeble W, Laursen S B, Poulsen K, Willis A C, Eggleton P, Hansen S, Holmskov U, Reid K B M, Jensenius J C. Nature (London) 1997;386:506–510. doi: 10.1038/386506a0. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doolittle R F. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 25.Arlaud G J, Gagnon J. Biosci Rep. 1981;1:779–784. doi: 10.1007/BF01114800. [DOI] [PubMed] [Google Scholar]
- 26.Brenner S. Nature (London) 1988;334:528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Kraut J. Annu Rev Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- 29.Ogata R T, Low P J, Kawakami M. J Immunol. 1995;154:2351–2357. [PubMed] [Google Scholar]
- 30.Matsushita M, Fujita T. Immunobiology. 1995;194:443–448. doi: 10.1016/S0171-2985(11)80110-5. [DOI] [PubMed] [Google Scholar]