Abstract
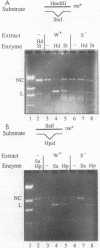
The methyl-directed DNA repair efficiency of a series of M13mp9 frameshift heteroduplexes 1, 2, or 3 unpaired bases was determined by using an in vitro DNA mismatch repair assay. Repair of hemimethylated frameshift heteroduplexes in vitro was directed to the unmethylated strand; was dependent on MutH, MutL, and MutS; and was equally efficient on base insertions and deletions. However, fully methylated frameshift heteroduplexes were resistant to repair, while totally unmethylated substrates were repaired with no strand bias. Hemimethylated 1-, 2-, or 3-base insertion and deletion heteroduplexes were repaired by the methyl-directed mismatch repair pathway as efficiently as the G.T mismatch. These results are consistent with earlier in vivo studies and demonstrate the involvement of methyl-directed DNA repair in the efficient prevention of frameshift mutations.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Au K. G., Cabrera M., Miller J. H., Modrich P. Escherichia coli mutY gene product is required for specific A-G----C.G mismatch correction. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9163–9166. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J., Krämmer G., Knippers R. Asymmetric repair of bacteriophage T7 heteroduplex DNA. Mol Gen Genet. 1981;181(4):541–547. doi: 10.1007/BF00428750. [DOI] [PubMed] [Google Scholar]
- Brown T., Hunter W. N., Kneale G., Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Kennard O., Kneale G., Rabinovich D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature. 1985 Jun 13;315(6020):604–606. doi: 10.1038/315604a0. [DOI] [PubMed] [Google Scholar]
- Bruni R., Martin D., Jiricny J. d(GATC) sequences influence Escherichia coli mismatch repair in a distance-dependent manner from positions both upstream and downstream of the mismatch. Nucleic Acids Res. 1988 Jun 10;16(11):4875–4890. doi: 10.1093/nar/16.11.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M., Nghiem Y., Miller J. H. mutM, a second mutator locus in Escherichia coli that generates G.C----T.A transversions. J Bacteriol. 1988 Nov;170(11):5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway M., Youderian P., Marinus M. G. Spontaneous mutations occur near dam recognition sites in a dam- Escherichia coli host. Genetics. 1987 Jul;116(3):343–347. doi: 10.1093/genetics/116.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy H. E., Fowler R. G. The specificity of base-pair substitution induced by the mutL and mutS mutators in E. coli. Mutat Res. 1985 Mar;142(3):93–97. doi: 10.1016/0165-7992(85)90046-6. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Dzidić S., Wagner R., Radman M. Large non-homology in heteroduplex DNA is processed differently than single base pair mismatches. Mol Gen Genet. 1987 Jan;206(1):181–184. doi: 10.1007/BF00326556. [DOI] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Methyl-directed repair of frameshift mutations in heteroduplex DNA. Proc Natl Acad Sci U S A. 1986 May;83(10):3395–3397. doi: 10.1073/pnas.83.10.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., Kolodner R. An Escherichia coli cell-free system that catalyzes the repair of symmetrically methylated heteroduplex DNA. Cold Spring Harb Symp Quant Biol. 1984;49:603–609. doi: 10.1101/sqb.1984.049.01.068. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., Siegel E. C., Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J Mol Biol. 1986 Mar 20;188(2):147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B., van den Elsen P., Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978 Jul 25;163(3):307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Hoess R. H. Cloning of mutH and identification of the gene product. Gene. 1983 May-Jun;22(2-3):245–253. doi: 10.1016/0378-1119(83)90109-9. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Hoess R. H. Nucleotide sequence of the Escherichia coli mutH gene. Nucleic Acids Res. 1987 Apr 10;15(7):3073–3084. doi: 10.1093/nar/15.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilley M., Welsh K. M., Su S. S., Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989 Jan 15;264(2):1000–1004. [PubMed] [Google Scholar]
- Haber L. T., Pang P. P., Sobell D. I., Mankovich J. A., Walker G. C. Nucleotide sequence of the Salmonella typhimurium mutS gene required for mismatch repair: homology of MutS and HexA of Streptococcus pneumoniae. J Bacteriol. 1988 Jan;170(1):197–202. doi: 10.1128/jb.170.1.197-202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Fan D. P. Further characterization of a non-essential mutator gene in Escherichia coli K-12. J Bacteriol. 1975 Nov;124(2):650–660. doi: 10.1128/jb.124.2.650-660.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Herman R. K. Isolation and characterization of mutator strains of Escherichia coli K-12. J Bacteriol. 1975 May;122(2):474–484. doi: 10.1128/jb.122.2.474-484.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Anand N. N., Kennard O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature. 1986 Apr 10;320(6062):552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- Jiricny J., Su S. S., Wood S. G., Modrich P. Mismatch-containing oligonucleotide duplexes bound by the E. coli mutS-encoded protein. Nucleic Acids Res. 1988 Aug 25;16(16):7843–7853. doi: 10.1093/nar/16.16.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua-Tor L., Rabinovich D., Hope H., Frolow F., Appella E., Sussman J. L. The three-dimensional structure of a DNA duplex containing looped-out bases. Nature. 1988 Jul 7;334(6177):82–84. doi: 10.1038/334082a0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Knowles J. R. A simple and efficient method for chemical mutagenesis of DNA. Nucleic Acids Res. 1985 Mar 11;13(5):1733–1745. doi: 10.1093/nar/13.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kramer W., Schughart K., Fritz H. J. Directed mutagenesis of DNA cloned in filamentous phage: influence of hemimethylated GATC sites on marker recovery from restriction fragments. Nucleic Acids Res. 1982 Oct 25;10(20):6475–6485. doi: 10.1093/nar/10.20.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem. 1986 Oct 15;261(29):13581–13587. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lahue R. S., Au K. G., Modrich P. DNA mismatch correction in a defined system. Science. 1989 Jul 14;245(4914):160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lahue R. S., Su S. S., Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong P. M., Hsia H. C., Miller J. H. Analysis of spontaneous base substitutions generated in mismatch-repair-deficient strains of Escherichia coli. J Bacteriol. 1986 Oct;168(1):412–416. doi: 10.1128/jb.168.1.412-416.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene S. D., Zimm B. H. Separations of open-circular DNA using pulsed-field electrophoresis. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4054–4057. doi: 10.1073/pnas.84.12.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell. 1988 Sep 9;54(6):805–812. doi: 10.1016/s0092-8674(88)91109-9. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L. Influence of GATC sequences on Escherichia coli DNA mismatch repair in vitro. J Bacteriol. 1987 Mar;169(3):1254–1259. doi: 10.1128/jb.169.3.1254-1259.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längle-Rouault F., Maenhaut-Michel G., Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 1987 Apr;6(4):1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Harrison R. W., Wlodawer A., Appella E., Sussman J. L. Crystal structure of 15-mer DNA duplex containing unpaired bases. Nature. 1988 Jul 7;334(6177):85–86. doi: 10.1038/334085a0. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Olivera B. M., Lehman I. R. Diphosphopyridine nucleotide: a cofactor for the polynucleotide-joining enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1700–1704. doi: 10.1073/pnas.57.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- SALIVAR W. O., TZAGOLOFF H., PRATT D. SOME PHYSICAL-CHEMICAL AND BIOLOGICAL PROPERTIES OF THE ROD-SHAPED COLIPHAGE M13. Virology. 1964 Nov;24:359–371. doi: 10.1016/0042-6822(64)90173-4. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986 May 20;189(2):273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L. Escherichia coli mutT mutator effect during in vitro DNA synthesis. Enhanced A.G replicational errors. J Biol Chem. 1987 Dec 5;262(34):16267–16270. [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel E. C., Kamel F. Reversion of frameshift mutations by mutator genes in Escherichia coli. J Bacteriol. 1974 Mar;117(3):994–1001. doi: 10.1128/jb.117.3.994-1001.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh K. M., Lu A. L., Clark S., Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987 Nov 15;262(32):15624–15629. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]