Abstract
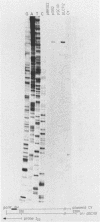
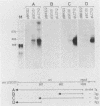
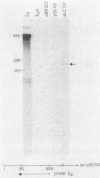
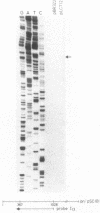
Insertion mutations were isolated in the origin fragment of the plasmid pSC101 after random cleavage with DNase I. The replication properties of the resulting plasmids confirmed previous findings and extended the characterization of the essential regions. Using these plasmids, we analyzed by various methods the transcription events in the pSC101 origin. In addition to the mRNA of repA, a gene coding for the self-regulated RepA protein which is essential for replication of the plasmid, we characterized a transcript, which we called RNA Y, that runs in the opposite direction and that starts in the middle of the second repeated sequence in the origin region. Like the self-regulated repA mRNA, RNA Y is weakly expressed. It does not code for a complete protein within the origin fragment but may do so in the wild-type plasmid. We also found indications for one or, possibly, two small RNA species, called RNA X, which run in the same direction as RepA and which are partially complementary to RNA Y. We postulate that RNA Y and, possibly, RNA X are implicated in the initiation of replication of pSC101.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Baker T. A., Funnell B. E., Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987 May 15;262(14):6877–6885. [PubMed] [Google Scholar]
- Bernardi A., Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984 Dec 21;12(24):9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: characterization of pSC101 mutants that replicate in the absence of IHF. J Bacteriol. 1989 Apr;171(4):2056–2065. doi: 10.1128/jb.171.4.2056-2065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Churchward G., Linder P., Caro L. The nucleotide sequence of replication and maintenance functions encoded by plasmid pSC101. Nucleic Acids Res. 1983 Aug 25;11(16):5645–5659. doi: 10.1093/nar/11.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Recircularization and autonomous replication of a sheared R-factor DNA segment in Escherichia coli transformants. Proc Natl Acad Sci U S A. 1973 May;70(5):1293–1297. doi: 10.1073/pnas.70.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977 Nov;132(2):734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Willetts N. S. Mobilization of the non-conjugative plasmid RSF1010: a genetic analysis of its origin of transfer. Mol Gen Genet. 1987 Jan;206(1):154–160. doi: 10.1007/BF00326551. [DOI] [PubMed] [Google Scholar]
- Frey J., Chandler M., Caro L. The effects of an Escherichia coli dnaAts mutation on the replication of the plasmids colE1 pSC101, R100.1 and RTF-TC. Mol Gen Genet. 1979 Jul 13;174(2):117–126. doi: 10.1007/BF00268349. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kaguni J. M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., Burger A. C., Churchward G., Caro L., Galas D., Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol Gen Genet. 1986 Jul;204(1):85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Effect of dna mutations on the replication of plasmid pSC101 in Escherichia coli K-12. J Bacteriol. 1979 Mar;137(3):1095–1099. doi: 10.1128/jb.137.3.1095-1099.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y., Ryter A., Jacob F. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb Symp Quant Biol. 1968;33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandjian E. W. UV crosslinking of RNA to nylon membrane enhances hybridization signals. Mol Biol Rep. 1986;11(2):107–115. doi: 10.1007/BF00364822. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lark K. G. Evidence for the direct involvement of RNA in the initiation of DNA replication in Escherichia coli 15T. J Mol Biol. 1972 Feb 28;64(1):47–60. doi: 10.1016/0022-2836(72)90320-8. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Caro L. Plasmid pSC101 replication mutants generated by insertion of the transposon Tn1000. J Mol Biol. 1983 Oct 25;170(2):287–303. doi: 10.1016/s0022-2836(83)80149-1. [DOI] [PubMed] [Google Scholar]
- Linder P., Churchward G., Xia G. X., Yu Y. Y., Caro L. An essential replication gene, repA, of plasmid pSC101 is autoregulated. J Mol Biol. 1985 Feb 5;181(3):383–393. doi: 10.1016/0022-2836(85)90227-x. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Melgar E., Goldthwait D. A. Deoxyribonucleic acid nucleases. II. The effects of metals on the mechanism of action of deoxyribonuclease I. J Biol Chem. 1968 Sep 10;243(17):4409–4416. [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W., Seufert W., Schaefer C., Gielow A., Hartmann H., Wende M. Functions of the DnaA protein of Escherichia coli in replication and transcription. Biochim Biophys Acta. 1988 Dec 20;951(2-3):351–358. doi: 10.1016/0167-4781(88)90106-6. [DOI] [PubMed] [Google Scholar]
- Miller C. A., Tucker W. T., Meacock P. A., Gustafsson P., Cohen S. N. Nucleotide sequence of the partition locus of Escherichia coli plasmid pSC101. Gene. 1983 Oct;24(2-3):309–315. doi: 10.1016/0378-1119(83)90091-4. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Hashimoto-Gotoh T., Timmis K. N. Location of two relaxation nick sites in R6K and single sites in pSC101 and RSF1010 close to origins of vegetative replication: implication for conjugal transfer of plasmid deoxyribonucleic acid. J Bacteriol. 1980 Dec;144(3):923–932. doi: 10.1128/jb.144.3.923-932.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P., Wickner W. Genetic mapping of the Escherichia coli leader (signal) peptidase gene (lep): a new approach for determining the map position of a cloned gene. J Bacteriol. 1983 May;154(2):569–572. doi: 10.1128/jb.154.2.569-572.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell. 1986 Oct 10;47(1):89–97. doi: 10.1016/0092-8674(86)90369-7. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983 Dec;35(2 Pt 1):495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. Primary structure of the essential replicon of the plasmid pSC101. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke C., Bastia D. The replication initiator protein of plasmid pSC101 is a transcriptional repressor of its own cistron. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2252–2256. doi: 10.1073/pnas.82.8.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Masamune Y. Autogenous regulation of synthesis of the replication protein in plasmid pSC101. Mol Gen Genet. 1985;200(3):362–367. doi: 10.1007/BF00425718. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Yamaguchi M. The replication origin of pSC101: the nucleotide sequence and replication functions of the ori region. Gene. 1984 Jul-Aug;29(1-2):211–219. doi: 10.1016/0378-1119(84)90181-1. [DOI] [PubMed] [Google Scholar]