Abstract
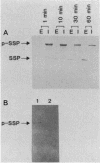
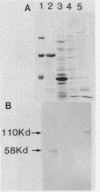
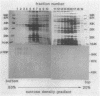
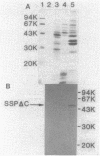
The Serratia marcescens serine protease, which is directed by the gene encoding a precursor composed of a typical NH2-terminal signal sequence, a mature enzyme domain, and a large COOH-terminal domain, was excreted through the outer membrane of Escherichia coli. The precursor, with the expected molecular size (110 kilodaltons), was detected in an insoluble form in the periplasmic space of E. coli cells after induction with isopropyl-beta-D-thiogalactopyranoside of the expression of the gene under the control of the tac promoter. Upon membrane fractionation of the disrupted cells by sucrose density gradient centrifugation, the precursor was recovered from a fraction slightly heavier than the outer membrane fraction but not from the inner membrane fraction. Conversion of the precursor into the mature form, which was accompanied by its excretion into the medium, was observed even in the absence of de novo protein synthesis caused by the addition of chloramphenicol. The mutated gene product lacking all of the COOH-terminal domain was localized in the periplasmic space only and was not excreted into the medium. Additional mutant genes were generated by site-directed mutagenesis to test the role of some amino acids in the excretion of this protease in E. coli. The mutant protein with no protease activity because of the change of the catalytic residue Ser-341 to Thr was still excreted into the medium but with abnormal processing. Both self-processing and host-dependent processing of the precursor seem to be involved in the excretion of the mature enzyme. Replacement of the four Cys residues, two in the mature enzyme and two in the COOH-terminal domain, with Ser in different combinations caused a distinct or complete loss of excretion, suggesting that a certain conformation possibly formed via disulfide bonding was important for the excretion of the S. marcescens protease.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Cornelis P., Digneffe C., Willemot K. Cloning and expression of a Bacillus coagulans amylase gene in Escherichia coli. Mol Gen Genet. 1982;186(4):507–511. doi: 10.1007/BF00337957. [DOI] [PubMed] [Google Scholar]
- Felmlee T., Pellett S., Lee E. Y., Welch R. A. Escherichia coli hemolysin is released extracellularly without cleavage of a signal peptide. J Bacteriol. 1985 Jul;163(1):88–93. doi: 10.1128/jb.163.1.88-93.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felmlee T., Pellett S., Welch R. A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985 Jul;163(1):94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L., Mackman N., Nicaud J. M., Holland I. B. The carboxy-terminal region of haemolysin 2001 is required for secretion of the toxin from Escherichia coli. Mol Gen Genet. 1986 Oct;205(1):127–133. doi: 10.1007/BF02428042. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakes K. S., Model P. Mechanism of export of colicin E1 and colicin E3. J Bacteriol. 1979 Jun;138(3):770–778. doi: 10.1128/jb.138.3.770-778.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Drutsa V., Jansen H. W., Kramer B., Pflugfelder M., Fritz H. J. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984 Dec 21;12(24):9441–9456. doi: 10.1093/nar/12.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Minsky A., Summers R. G., Knowles J. R. Secretion of beta-lactamase into the periplasm of Escherichia coli: evidence for a distinct release step associated with a conformational change. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4180–4184. doi: 10.1073/pnas.83.12.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T., Mizushima S. Separation and properties of outer and cytoplasmic membranes in Escherichia coli. Biochim Biophys Acta. 1969;193(2):268–276. doi: 10.1016/0005-2736(69)90188-6. [DOI] [PubMed] [Google Scholar]
- Pohlner J., Halter R., Beyreuther K., Meyer T. F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. 1987 Jan 29-Feb 4Nature. 325(6103):458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D. H. Precolicin E1, the major gene product of plasmid-ColE1 deoxyribonucleic acid in vitro. Biochem J. 1980 Feb 1;185(2):463–471. doi: 10.1042/bj1850463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida N., Uozumi T., Beppu T. Specific excretion of Serratia marcescens protease through the outer membrane of Escherichia coli. J Bacteriol. 1986 Jun;166(3):937–944. doi: 10.1128/jb.166.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]