Abstract
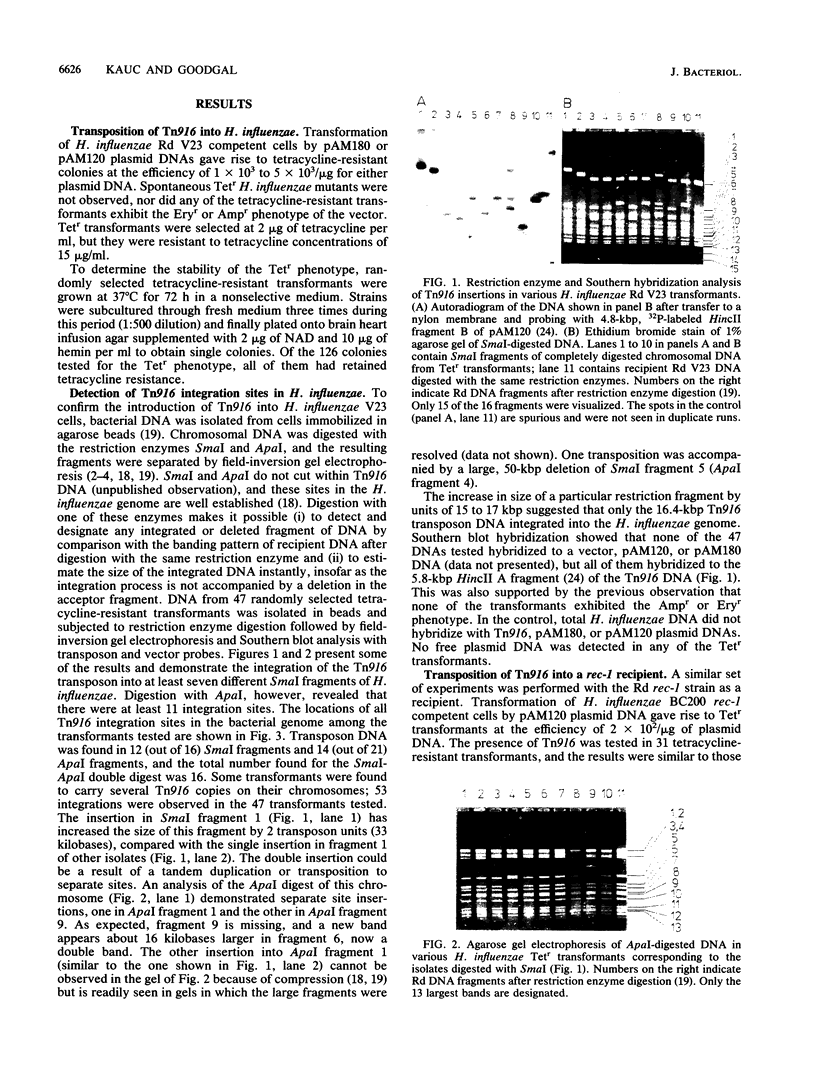
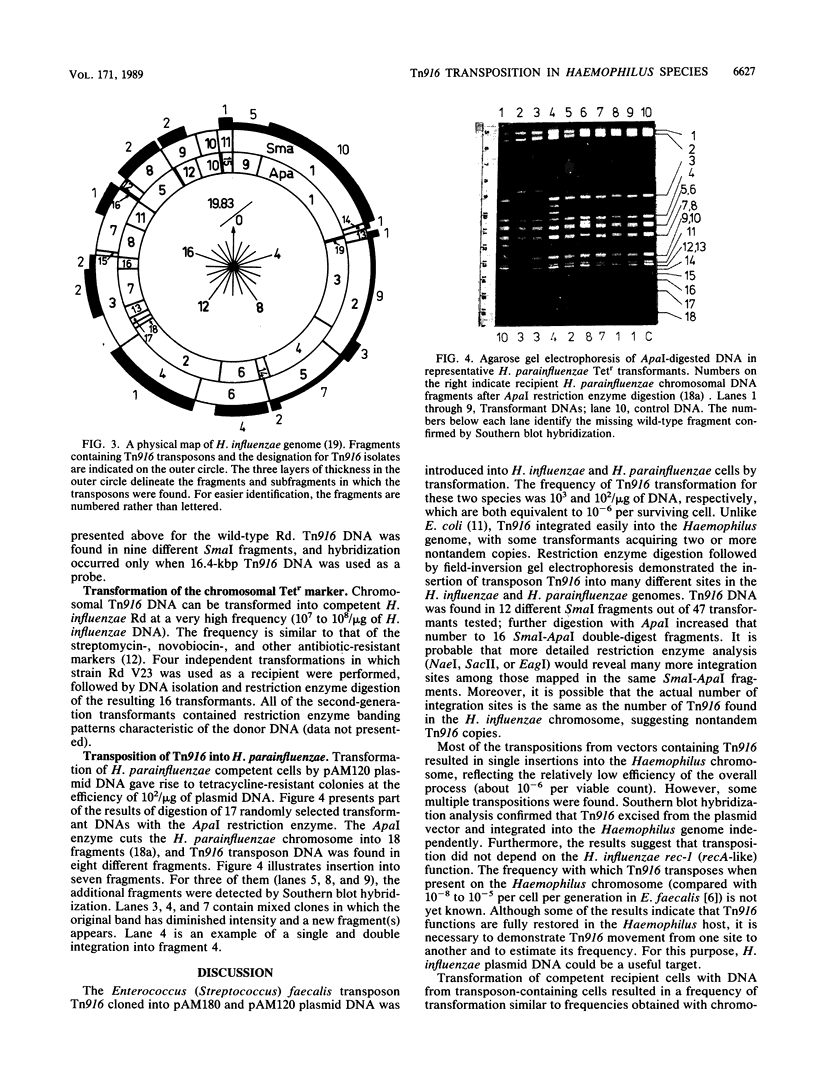
Enterococcus (Streptococcus) faecalis transposon Tn916 was introduced into Haemophilus influenzae Rd and Haemophilus parainfluenzae by transformation and demonstrated to transpose efficiently. Haemophilus transformants resistant to tetracycline were observed at a frequency of approximately 3 x 10(2) to 5 x 10(3)/micrograms of either pAM120 (pGL101::Tn916) or pAM180 (pAM81::Tn916) plasmid DNAs, which are incapable of autonomous replication in this host. Restriction enzyme analysis and Southern blot hybridization revealed that (i) Tn916 integrates into many different sites in the H. influenzae and H. parainfluenzae genomes; (ii) only the 16.4-kilobase-pair Tn916 DNA integrates, and no vector DNA was detected; and (iii) the Tetr phenotype was stable in the absence of selective pressure. Second-generation Tn916 transformants occurred at the high frequency of chromosomal markers and retained their original chromosomal locations. Similar results were obtained with H. influenzae Rd BC200 rec-1 as the recipient strain, which suggests host rec functions are not required in Tn916 integrative transposition. Transposition with Tn916 is an important procedure for mutagenesis of Haemophilus species.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock C. J. Parameters of field inversion gel electrophoresis for the analysis of pox virus genomes. Nucleic Acids Res. 1988 May 25;16(10):4239–4252. doi: 10.1093/nar/16.10.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R., Smith C. L., Mathew M. K. Pulsed-field gel electrophoresis of very large DNA molecules. Annu Rev Biophys Biophys Chem. 1988;17:287–304. doi: 10.1146/annurev.bb.17.060188.001443. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Korman R. Z., Zahler S. A., Adsit J. C., Dunny G. M. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J Bacteriol. 1987 Jun;169(6):2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- Deich R. A., Green B. A. Mobilization of Haemophilus influenzae chromosomal markers by an Escherichia coli F' factor. J Bacteriol. 1987 May;169(5):1905–1910. doi: 10.1128/jb.169.5.1905-1910.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Gerardot C. J. Use of pulsed-field-gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene. 1988 Sep 7;68(2):323–333. doi: 10.1016/0378-1119(88)90035-2. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982 Nov 18;300(5889):281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984 Jul;159(1):214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodgal S. H. DNA uptake in Haemophilus transformation. Annu Rev Genet. 1982;16:169–192. doi: 10.1146/annurev.ge.16.120182.001125. [DOI] [PubMed] [Google Scholar]
- Gromkova R., Goodgal S. Transformation by plasmid and chromosomal DNAs in Haemophilus parainfluenzae. Biochem Biophys Res Commun. 1979 Jun 27;88(4):1428–1434. doi: 10.1016/0006-291x(79)91139-2. [DOI] [PubMed] [Google Scholar]
- Herriott R. M., Meyer E. M., Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970 Feb;101(2):517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn G., Laufs R., Kaulfers P. M., Kolenda H. Molecular nature of two Haemophilus influenzae R factors containing resistances and the multiple integration of drug resistance transposons. J Bacteriol. 1979 May;138(2):584–597. doi: 10.1128/jb.138.2.584-597.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. M., Yost S. C., Pattee P. A. Transfer of the conjugal tetracycline resistance transposon Tn916 from Streptococcus faecalis to Staphylococcus aureus and identification of some insertion sites in the staphylococcal chromosome. J Bacteriol. 1987 May;169(5):2121–2131. doi: 10.1128/jb.169.5.2121-2131.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathariou S., Metz P., Hof H., Goebel W. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J Bacteriol. 1987 Mar;169(3):1291–1297. doi: 10.1128/jb.169.3.1291-1297.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauc L., Goodgal S. H. Amplification of DNA at a prophage attachment site in Haemophilus influenzae. J Bacteriol. 1989 Apr;171(4):1898–1903. doi: 10.1128/jb.171.4.1898-1903.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauc L., Goodgal S. H. The size and a physical map of the chromosome of Haemophilus parainfluenzae. Gene. 1989 Nov 30;83(2):377–380. doi: 10.1016/0378-1119(89)90125-x. [DOI] [PubMed] [Google Scholar]
- Kauc L., Mitchell M., Goodgal S. H. Size and physical map of the chromosome of Haemophilus influenzae. J Bacteriol. 1989 May;171(5):2474–2479. doi: 10.1128/jb.171.5.2474-2479.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B., Buu-Hoi A., Marshall B. Transposon Tn10-like tetracycline resistance determinants in Haemophilus parainfluenzae. J Bacteriol. 1984 Oct;160(1):87–94. doi: 10.1128/jb.160.1.87-94.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICKEL L., GOODGAL S. H. EFFECT OF INTERSPECIFIC TRANSFORMATION ON LINKAGE RELATIONSHIPS OF MARKERS IN HAEMOPHILUS INFLUENZAE AND HAEMOPHILUS PARAINFLUENZAE. J Bacteriol. 1964 Dec;88:1538–1544. doi: 10.1128/jb.88.6.1538-1544.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglich J. G., Andrews R. E., Jr Introduction of the Streptococcus faecalis transposon Tn916 into Bacillus thuringiensis subsp. israelensis. Plasmid. 1988 Mar;19(2):84–93. doi: 10.1016/0147-619x(88)90047-9. [DOI] [PubMed] [Google Scholar]
- Senghas E., Jones J. M., Yamamoto M., Gawron-Burke C., Clewell D. B. Genetic organization of the bacterial conjugative transposon Tn916. J Bacteriol. 1988 Jan;170(1):245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Boling M. E., Beattie K. L., Kimball R. F. A complex of recombination and repair genes in Haemophilus influenzae. J Mol Biol. 1972 Jul 21;68(2):361–378. doi: 10.1016/0022-2836(72)90218-5. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]






