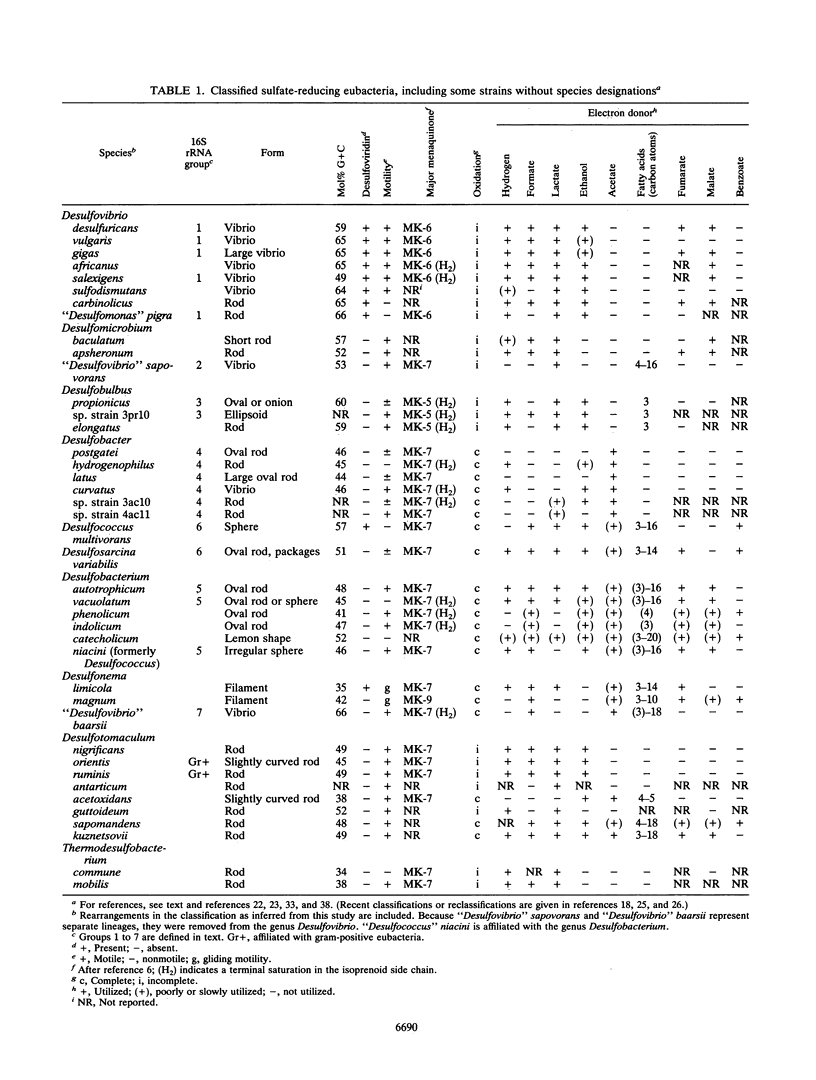
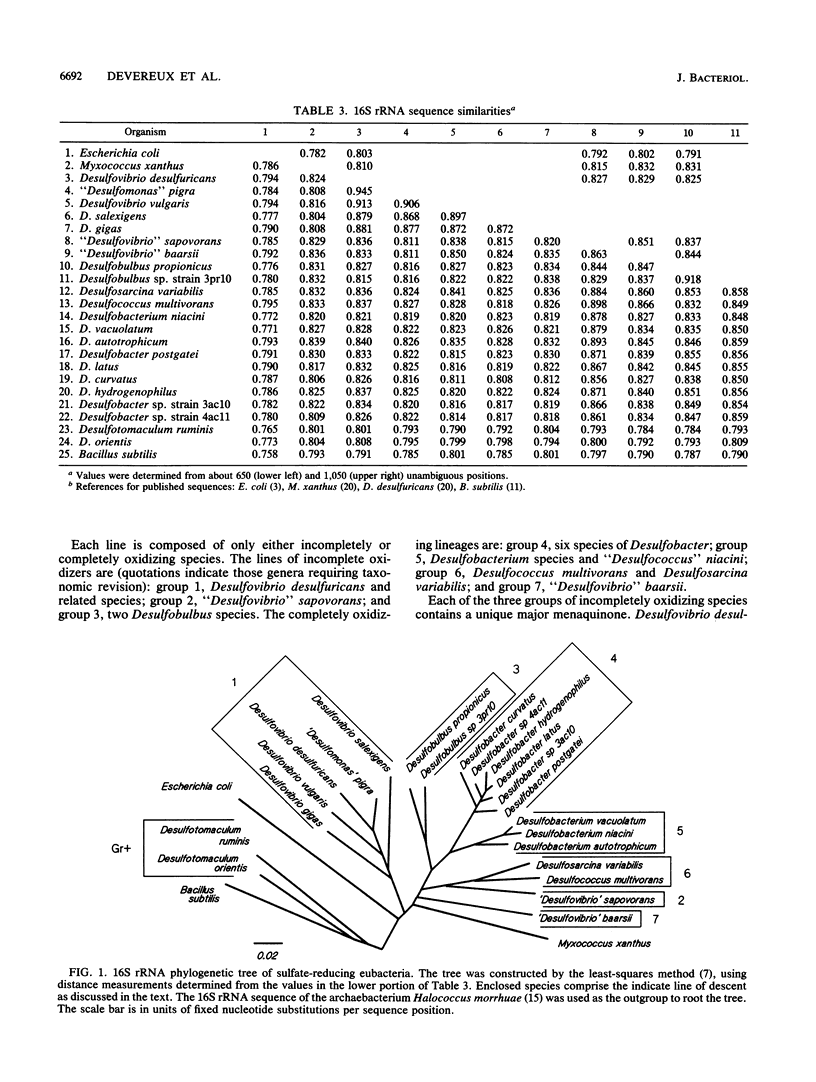
Abstract
Phylogenetic relationships among 20 nonsporeforming and two endospore-forming species of sulfate-reducing eubacteria were inferred from comparative 16S rRNA sequencing. All genera of mesophilic sulfate-reducing eubacteria except the new genus Desulfomicrobium and the gliding Desulfonema species were included. The sporeforming species Desulfotomaculum ruminis and Desulfotomaculum orientis were found to be gram-positive organisms sharing 83% 16S rRNA sequence similarity, indicating that this genus is diverse. The gram-negative nonsporeforming species could be divided into seven natural groups: group 1, Desulfovibrio desulfuricans and other species of this genus that do not degrade fatty acids (this group also included "Desulfomonas" pigra); group 2, the fatty acid-degrading "Desulfovibrio" sapovorans; group 3, Desulfobulbus species; group 4, Desulfobacter species; group 5, Desulfobacterium species and "Desulfococcus" niacini; group 6, Desulfococcus multivorans and Desulfosarcina variabilis; and group 7, the fatty acid-oxidizing "Desulfovibrio" baarsii. (The quotation marks are used to indicate the need for taxonomic revision.) Groups 1 to 3 are incomplete oxidizers that form acetate as an end product; groups 4 to 7 are complete oxidizers. The data were consistent with and refined relationships previously inferred by oligonucleotide catalogs of 16S rRNA. Although the determined relationships are generally consistent with the existing classification based on physiology and other characteristics, the need for some taxonomic revision is indicated.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achenbach-Richter L., Stetter K. O., Woese C. R. A possible biochemical missing link among archaebacteria. Nature. 1987 May 28;327(6120):348–349. doi: 10.1038/327348a0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Campbell L. L., Postgate J. R. Classification of the spore-forming sulfate-reducing bacteria. Bacteriol Rev. 1965 Sep;29(3):359–363. doi: 10.1128/br.29.3.359-363.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H., Garrett R. A. The nucleotide sequence of the 16S ribosomal RNA gene of the archaebacterium Halococcus morrhua. EMBO J. 1984 Jul;3(7):1613–1619. doi: 10.1002/j.1460-2075.1984.tb02019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Postgate J. R., Campbell L. L. Classification of Desulfovibrio species, the nonsporulating sulfate-reducing bacteria. Bacteriol Rev. 1966 Dec;30(4):732–738. doi: 10.1128/br.30.4.732-738.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter K. O., Lauerer G., Thomm M., Neuner A. Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science. 1987 May 15;236(4803):822–824. doi: 10.1126/science.236.4803.822. [DOI] [PubMed] [Google Scholar]
- Widdel F., Pfennig N. A new anaerobic, sporing, acetate-oxidizing, sulfate-reducing bacterium, Desulfotomaculum (emend.) acetoxidans. Arch Microbiol. 1977 Feb 4;112(1):119–122. doi: 10.1007/BF00446665. [DOI] [PubMed] [Google Scholar]
- Widdel F., Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch Microbiol. 1981 Jul;129(5):395–400. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]


