Abstract
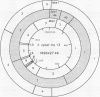
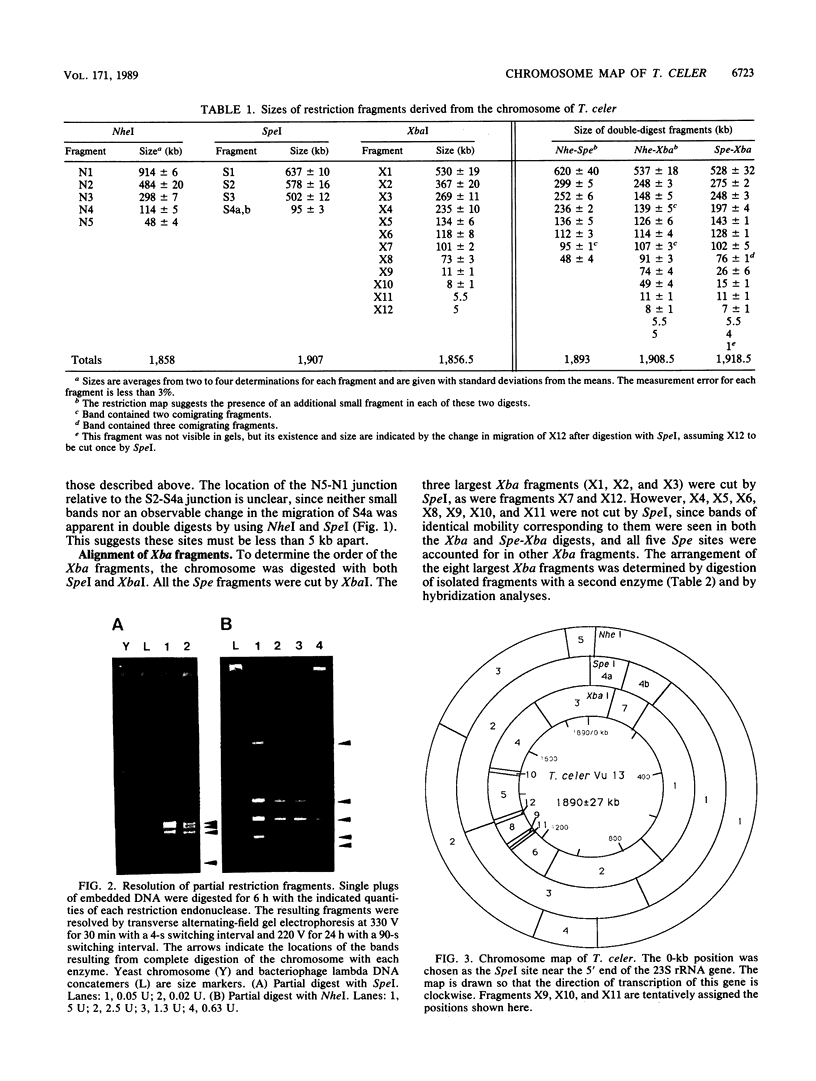
A physical map for the chromosome of the thermophilic archaebacterium Thermococcus celer Vu13 has been constructed. Thirty-four restriction endonucleases were tested for their ability to generate large restriction fragments from the chromosome of T. celer. Of these, the enzymes NheI, SpeI, and XbaI yielded the fewest fragments when analyzed by pulsed-field electrophoresis. NheI and SpeI each gave 5 fragments, while XbaI gave 12. The size of the T. celer chromosome was determined from the sum of the apparent sizes of restriction fragments derived from single and double digests by using these enzymes and was found to be 1,890 +/- 27 kilobase pairs. Partial and complete digests allowed the order of all but three small (less than 15 kilobase pairs) fragments to be deduced. These three fragments were assigned positions by using hybridization probes derived from these restriction fragments. The positions of the other fragments were confirmed by using hybridization probes derived in the same manner. The positions of the 5S, 16S, and 23S rRNA genes as well as the 7S RNA gene were located on this map by using cloned portions of these genes as hybridization probes. The 5S rRNA gene was localized 48 to 196 kilobases from the 5' end of the 16S gene. The 7S RNA gene was localized 190 to 504 kilobases from the 3' end of the 23S gene. These analyses demonstrated that the chromosome of T. celer is a single, circular DNA molecule. This is the first such demonstration of the structure of an archaebacterial chromosome.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achenbach-Richter L., Gupta R., Zillig W., Woese C. R. Rooting the archaebacterial tree: the pivotal role of Thermococcus celer in archaebacterial evolution. Syst Appl Microbiol. 1988;10:231–240. doi: 10.1016/s0723-2020(88)80007-9. [DOI] [PubMed] [Google Scholar]
- Achenbach-Richter L., Woese C. R. The ribosomal gene spacer region in archaebacteria. Syst Appl Microbiol. 1988;10:211–214. doi: 10.1016/s0723-2020(88)80002-x. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Daniels C. J., Reeve J. N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16(4):287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- Charlebois R. L., Hofman J. D., Schalkwyk L. C., Lam W. L., Doolittle W. F. Genome mapping in halobacteria. Can J Microbiol. 1989 Jan;35(1):21–29. doi: 10.1139/m89-004. [DOI] [PubMed] [Google Scholar]
- Culham D. E., Nazar R. N. An unlinked 5 S ribosomal RNA gene in the archaebacterium, Thermococcus celer. Mol Gen Genet. 1988 May;212(2):382–385. doi: 10.1007/BF00334712. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Laas W., Patterson D. Fractionation of large mammalian DNA restriction fragments using vertical pulsed-field gradient gel electrophoresis. Somat Cell Mol Genet. 1986 Mar;12(2):185–195. doi: 10.1007/BF01560665. [DOI] [PubMed] [Google Scholar]
- Kikuchi A., Asai K. Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984 Jun 21;309(5970):677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- Klein A., Schnorr M. Genome complexity of methanogenic bacteria. J Bacteriol. 1984 May;158(2):628–631. doi: 10.1128/jb.158.2.628-631.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meile L., Leisinger T., Reeve J. N. Cloning of DNA sequences from Methanococcus vannielii capable of autonomous replication in yeast. Arch Microbiol. 1985 Dec;143(3):253–255. doi: 10.1007/BF00411245. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Simon M. I., Barbour A. G. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985 Nov 21;318(6043):257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- Sapienza C., Doolittle W. F. Unusual physical organization of the Halobacterium genome. Nature. 1982 Feb 4;295(5848):384–389. doi: 10.1038/295384a0. [DOI] [PubMed] [Google Scholar]
- Stewart G., Furst A., Avdalovic N. Transverse alternating field electrophoresis (TAFE). Biotechniques. 1988 Jan;6(1):68–73. [PubMed] [Google Scholar]
- Waterbury P. G., Lane M. J. Generation of lambda phage concatemers for use as pulsed field electrophoresis size markers. Nucleic Acids Res. 1987 May 11;15(9):3930–3930. doi: 10.1093/nar/15.9.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]