Abstract
The molecular basis of the polymorphic tumor rejection antigens of chemically induced sarcomas of inbred mice remains a mystery, despite the discovery of these antigens over 40 years ago and their critical importance to the foundation of tumor immunology. In an analysis of a panel of BALB/c 3-methylcholanthrene-induced tumors, we identified one tumor, CMS5, that elicited a strong cytotoxic T cell response with exquisite specificity for CMS5. A stable cloned line of T cells with this specificity (C18) was used to screen a CMS5 cDNA expression library. The gene encoding the C18-defined antigen was identified as a mutated form of a mouse mitogen-activated protein kinase, ERK2, and a peptide incorporating the resulting amino acid substitution (lysine to glutamine) was efficiently recognized by C18. Vaccination with this peptide elicited specific resistance to CMS5 challenge. Extensive efforts to isolate antigen-loss variants of CMS5 were unsuccessful, suggesting that the mutated mitogen-activated protein kinase is essential for maintenance of the malignant phenotype.
The finding that sarcomas induced by cancer carcinogens are immunogenic in inbred strains of mice is one of the major milestones in the field of tumor immunology. Using various carcinogens and different inbred strains of mice, many investigators have confirmed the original observations of Gross (1), Foley (2), and Prehn and Main (3) that mice can be rendered resistant to tumor transplantation by preimmunization with the same tumor (either by previous growth and removal of the tumor or by injection of irradiated tumor cells) (4–6). Resistance does not generally extend to other syngeneic tumors induced by the same carcinogen, but is restricted to challenge with the tumor used for immunization, indicating that the antigenic polymorphism of chemically induced sarcomas is apparently quite extensive (7, 8). There have been many attempts to isolate and characterize the tumor antigens eliciting this transplantation immunity, but as yet, none have been defined.
Immunity to transplants of chemically induced sarcomas can be adoptively transferred to nonimmunized hosts by T cells from actively immunized donors, whereas passive transfer of serum from immunized donors is ineffective (9, 10). Attempts to analyze the in vitro specificity of T cells from immunized donors by cytotoxicity tests have not given clear-cut results. Although T cells with cytotoxicity for the immunizing tumor can be isolated from immunized donors, these T cells show a complex pattern of reactivity ranging from apparent specificity for the immunizing tumor to crossreactivity with other syngeneic sarcomas (11, 12). Because activation of murine leukemia virus is frequently associated with tumorgenesis in the mouse (13), murine leukemia virus-related antigens frequently are expressed by sarcomas, and their presence has complicated the interpretation of the in vitro specificity analysis of T cells from immunized mice.
We reinvestigated the reactivity of cytotoxic T lymphocytes (CTL) from donors immunized against BALB/c 3-methylcholanthrene (3-MC)-induced sarcomas and detected particularly strong cytotoxicity against the BALB/c sarcoma, CMS5. Using a CMS5-specific CTL clone to screen a cDNA library, we identified a mutated form of mitogen-activated protein kinase as the source of the CMS5-specific antigen.
MATERIALS AND METHODS
Tumors.
The tumors listed in Fig. 1A have been described previously (5, 14, 15). CMS5a and CMS5j are cloned tumor lines obtained from CMS5 by limiting dilution. The tumor necrosis factor (TNF)-sensitive WEHI164S line was provided by E. Lattime (Thomas Jefferson University) (16). P1. HTR is a subline derived from P815 mastocytoma of DBA/2 origin (17).
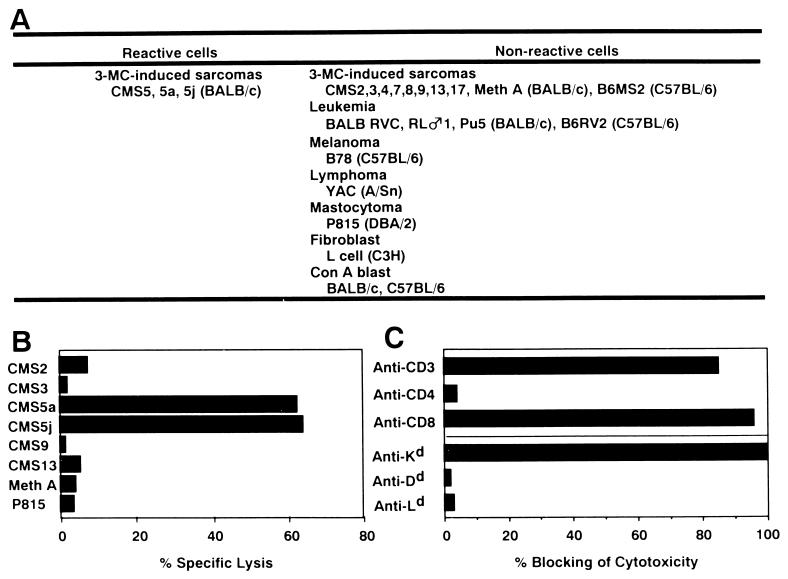
Figure 1.
CTL clone C18 recognizes an antigen expressed by CMS5 with H-2Kd restriction. C18 displays high lytic activity on CMS5 and its two sublines CMS5a and CMS5j, but does not lyse other cell types, including nine syngeneic 3-MC-induced fibrosarcomas. (A) Summary of 51Cr-release cytotoxicity assays with C18. (B) Example of a typical cytotoxicity assay. (C) Lytic activity of C18 against CMS5a was strongly inhibited by addition of mAbs against CD3, CD8, and Kd, but not by the mAbs against CD4, Dd, or Ld.
Antibodies.
Anti-CD3 mAb, produced by hybridoma 145-2C11 (18), anti-L3T4 (CD4) mAb, produced by hybridoma GK1.5 (19), anti-Lyt-2.2 (CD8) mAb, produced by hybridoma 19/178 (20), anti-H-2Kd mAb, produced by hybridoma 20-8-4 (21), anti-H-2Ld mAb, produced by hybridoma 30-5-7 (22), and anti-H-2Dd mAb, produced by hybridoma 34-5-8S (23) were used for blocking of CTL activity in 51Cr-release cytotoxicity assay.
Recombinant Murine Interleukin 12 (IL-12).
Recombinant murine IL-12 (24, 25) was provided by Genetics Institute (Cambridge, MA).
Establishment of CTL Clones.
Eight-week-old BALB/c female mice were inoculated subcutaneously with 2.5 × 105 CMS5a cells. Nine days later, 2 × 106 inguinal lymph node cells were obtained and cultured with 1 × 105 cells of irradiated (100 Gy) CMS5a cells and 1 × 105 irradiated (30 Gy) syngeneic peritoneal exudate cells in 24-well culture plates. After 7–14 days, the cultured cells were harvested, and limited numbers (1 to 20) were cultured with 5 × 103 irradiated (100 Gy) CMS5a cells and 5 × 103 irradiated (30 Gy) syngeneic peritoneal exudate cells in 96-well culture plates. After initial expansion of clones in 96-well plates, the cells were recloned at 0.8 cells/well by limiting dilution, and a CTL clone designated C18 was established. CTL were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 50 μM 2-mercaptoethanol, 20 units/ml recombinant IL-12 at 37°C in a 5% CO2 atmosphere.
51Cr-Release Cytotoxicity Assay.
The assay was performed as described (26). Briefly, target cells (2 × 106) were labeled with 1.85 MBq of Na251CrO4 in 0.2 ml of culture medium for 1 hr at 37°C under 5% CO2 in air. The labeled target cells (1 × 104) were incubated with 5 × 104 C18 cells for 6 hr at 37°C. For blocking CTL activity with mAbs, serially diluted mAbs were added to C18 and labeled target cells before the cytotoxicity assay.
TNF Release Assay.
The TNF release assay was performed as described (27). Briefly, 2–4 × 103 C18 cells were mixed with 3 × 104 target cells in 96-well plates for 24 hr at 37°C under 5% CO2 in air. Fifty microliters of supernatant collected from each well was added to 4 × 103 TNF-sensitive WEHI164S cells in 50 μl of RPMI 1640 medium with 10% fetal calf serum in flat-bottom 96-well plates. Plates were incubated for 48 hr at 37°C under 5% CO2, and wells were pulsed with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (thiazolyl blue) at a concentration of 0.4 μg/ml for 4 hr, and cells were dissolved in 150 μl of isopropyl alcohol containing 0.04 M HCl and 0.1% Nonidet P-40. Absorbance was measured at 570 nm.
Construction of the cDNA Library.
Total RNA was isolated from CMS5j and poly(A)+ RNA was prepared by oligo(dT)-cellulose column (mRNA purification kit; Pharmacia). mRNA was converted to cDNA, ligated to BstXI adaptors, and inserted into the BstXI site of expression vector pCDM8 (Invitrogen) as described in the superscript Choice System (GIBCO/BRL).
DNA Sequencing and Homology Search.
DNA sequence analysis was performed by double-stranded sequencing using the Taq Dye Primer Cycle sequencing kit (Applied Biosystems) and a DNA sequencer (model 370A; Applied Biosystems). The computer search for sequence homology was performed by the program blast with GenBank databases.
Reverse Transcription–PCR and Sequence Analysis of PCR Product.
cDNA was synthesized from total RNAs of cell lines with an oligo(dT)14 primer. Thirty cycles of PCR (94°C, 1 min; 50°C, 1 min; 72°C, 2 min) were performed with 5′ primer CCAACCATTGAGCAAATGAA (from 271 to 290: position 1 = A of ATG) and 3′ primer CAAGGCCAAAGTCACAGATC (from 486 to 505) derived from the mouse ERK2 sequence. PCR products were purified with Microcon 100 (Amicon) and sequenced directly or after subcloning into the pCRII vector.
Preparation of Peptide-Pulsed Target Cells.
Peptides were purchased from Sawady Technology (Tokyo). One million P1. HTR cells were pulsed with peptides at various concentrations for 1 hr at room temperature and 1 hr at 37°C under 5% CO2, and used as target cells for the 51Cr-release cytotoxicity assay.
Competitive Inhibition Assay of Peptide Binding.
For competitive inhibition of peptide binding to Kd class I molecule, 51Cr-labeled P1. HTR cells were pulsed with the Kd-binding erbB2-derived peptide (TYLPTNASL) at 0.1 nM in the presence of serially diluted ERK2-derived peptides (9m, 9wt, and 3m). After washing, anti-erbB2 CTL line B3, which is known to recognize the peptide TYLPTNASL in the context of Kd, was added at an effector-to-target ratio of 5, and percentage-specific 51Cr-release was measured. P1. HTR cells pulsed with the erbB2-derived peptide in the absence of inhibitory peptides showed 80% B3-directed specific lysis.
RESULTS AND DISCUSSION
C18 Is a CTL Clone Specific for the MC-Induced Sarcoma CMS5.
CMS5 was induced in a female BALB/c mouse by subcutaneous injection of 0.1 mg of 3-MC (14). In transplantation studies, the tumor rejection antigen of CMS5 was strong and characteristically non-crossreactive with other 3-MC-induced BALB/c tumors. Mice injected with CMS5 developed CTL with specificity for CMS5, and a long-term CTL clone, C18, was established from the inguinal lymph node of a CMS5-immunized mouse and was maintained by stimulation with 100 Gy-irradiated CMS5 and 30 Gy-irradiated syngeneic peritoneal cells every 2 weeks.
Fig. 1A summarizes the specificity analysis of C18 in 51Cr-release cytotoxicity assays and Fig. 1B the result of an individual test. C18 reactivity is restricted to CMS5 and subclones derived from CMS5, e.g. CMS5a and j. C18 did not lyse nine other BALB/c 3-MC-induced sarcomas or a variety of other cell types. Blocking studies showed that C18 is a Kd-restricted CD8+ T cell clone (Fig. 1C). The TNF-release assay (27) showed the same specificity as the 51Cr-release cytotoxicity assay.
cDNA Clone 84-53-13 Codes for an Antigen Recognized by C18 in the Context of Kd.
For cloning (28) the gene responsible for the C18-defined antigen, a cDNA library from CMS5j was prepared, ligated to BstXI adapters, and inserted into the BstXI site of the expression vector pCDM8 (Invitrogen). The library was divided into pools of about 100 bacterial colonies, and 100 ng of DNA from each pool and 100 ng of plasmid pdl3027 containing the polyoma T antigen gene (29) were cotransfected by the DEAE-dextran/chloroquine method (30) into 2 × 104 CMS8 cells (a non-crossreactive BALB/c sarcoma). After 48 hr, 2 × 103 C18 cells were added, and 24 hr later the supernatants were assayed for TNF release. A single positive cDNA clone, 84-53-13, was isolated after screening 70,000 cDNA colonies. To prove that 84–53-13 encodes the C18-defined antigen, it was transiently transfected with or without the Kd restriction element into a variety of C18 nonreactive cell lines, including four BALB/c sarcomas by the DEAE-dextran-chloroquine method. The 84-53-13 transfected BALB/c cells became reactive with C18, whereas non-BALB/c cells became reactive only if 84-53-13 was cotransfected with Kd (Fig. 2A).
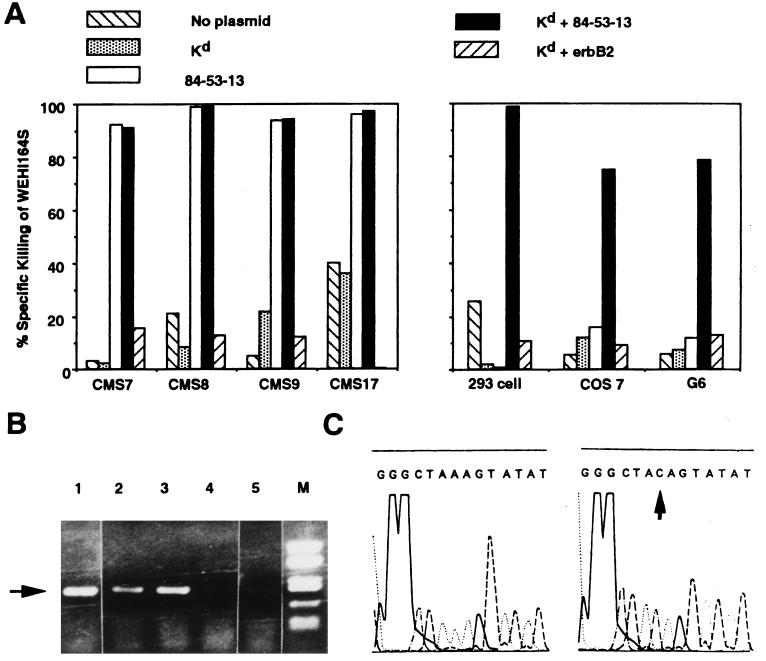
Figure 2.
The cDNA clone 84-53-13 encodes the CMS5 antigen recognized by C18. (A) Non-crossreactive BALB/c sarcomas CMS7, 8, 9, and 17 became C18 reactive in the TNF-release assay after transfection with 84-53-13. 293 cells (human embryonal kidney), COS 7 cells (monkey kidney), and G6 cells (melanoma line of C57BL/6 origin) required cotransfection with 84-53-13 and Kd to become C18 reactive. Transfection with a control plasmid (pCDM8) coding for erbB2 with Kd did not result in C18 reactivity. (B) Reverse transcription-PCR with primers for ERK2 were carried out with RNA obtained from CMS5j, CMS5a, CMS7, CMS8, and L cells (arrowhead in lanes 1–5, respectively). (C) PCR products from CMS5j were sequenced. Three of six clones carried the AAG → CAG mutation (Right), whereas the remaining clones had the wild-type sequence (Left).
Clone 84-53-13 Codes for Mitogen-Activated Protein Kinase ERK2 with an Amino Acid Substitution.
Clone 84-53-13 contained a cDNA insert of 1,498 bp whose ORF encoded a 358-amino acid peptide. The sequence of this clone was identical to ERK2 (31), a mouse mitogen-activated protein kinase, except for a single base pair resulting in an amino acid replacement at position 136 from a lysine to a glutamine. To exclude a cloning artifact, reverse transcription-PCR using primers derived from wild-type ERK2 were performed with RNA derived from CMS5j, CMS5a, CMS7, CMS8, and L cell (Fig. 2B). CMS7, CMS8, and L cell contained the wild-type AAG codon (lysine) at position 136. Sequences from CMS5a and CMS5j contained both the AAG (lysine) and mutated CAG (glutamine) codons at position 136 (arrowhead in Fig. 2C), confirming the initial 84–53-13 cloning and demonstrating that CMS5 also has a wild-type allele of ERK2.
A Peptide 9m Derived from Mutated ERK2 Presents a Novel Epitope Recognized by C18.
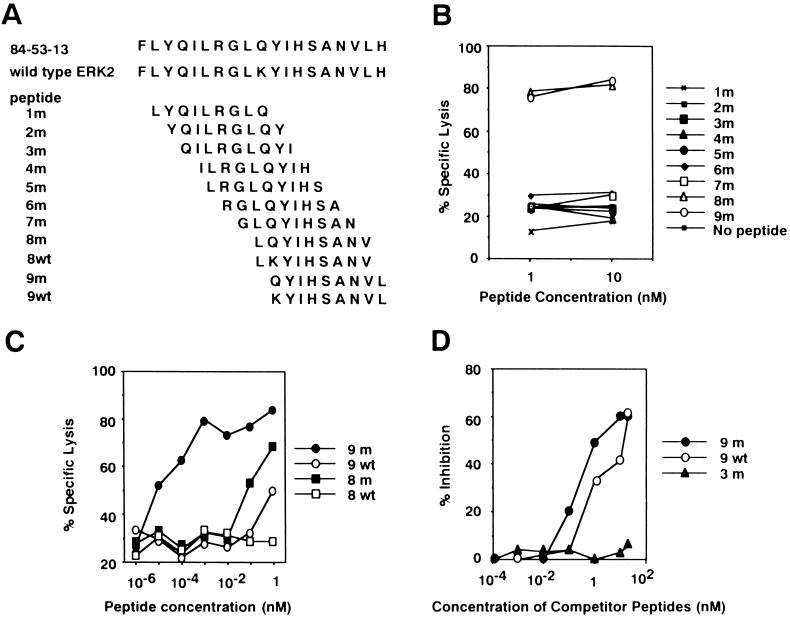
To determine the antigenic peptide recognized by C18, a series of synthetic peptides spanning position 136 were synthesized (Fig. 3A). The two peptides with the substituted amino acid, 8m and 9m, sensitized target cells equally well against C18 lysis at concentrations of 1 nM and 10 nM (Fig. 3B). When peptides were further diluted, 9m was more efficient than 8m in sensitization (Fig. 3C). The corresponding wild-type peptide, 9wt, showed marginal sensitization to C18 lysis at a concentration of 1 nM. The binding affinity of peptides 9m and 9wt to Kd was examined by measuring their ability to inhibit the sensitization of target cells to an unrelated Kd-binding peptide. The two peptides showed comparable inhibitory effects, whereas a control peptide, 3m, had no inhibitory activity (Fig. 3D). Taken together, the results indicate that the 9m peptide is efficiently recognized by C18, and that the lysine-to-glutamine substitution at position 136 generates a new epitope rather than a new agretope. Two additional CTL clones, C23.1 and C15.1, independently isolated from mice immunized with CMS5 also showed specificity for 9m peptide (data not shown).
Figure 3.
Identification of an antigenic peptide recognized by C18. (A) Serial 9-mer synthetic peptides were synthesized incorporating the position 136 amino acid in the mutated (1m to 9m) or nonmutated (8wt and 9wt) ERK2 product. (B) Two peptides, 8m and 9m, sensitized DBA/2 (H-2d)-derived P1. HTR target cells equally well against C18 lysis at concentrations of 1 nM and 10 nM. (C) The 9m peptide was more efficient in sensitizing P1. HTR target to C18 lysis than other peptides. (D) The affinity of peptides 9m and 9wt for Kd was measured in a binding-inhibition assay using a Kd-restricted human erbB2 peptide. Peptides 9m and 9wt had comparable Kd binding affinity, whereas the 3m peptide had no inhibitory activity.
The 9m Peptide Is a Tumor Rejection Antigen of CMS5.
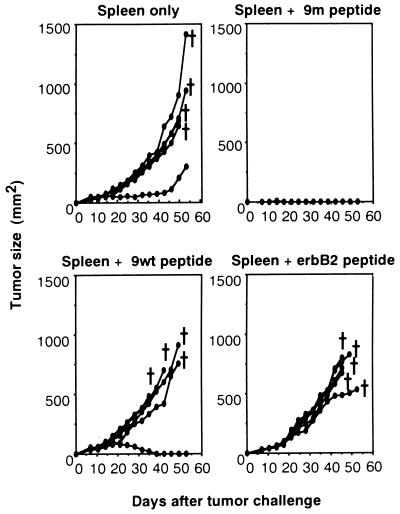
To determine whether the 9m peptide can function as a tumor rejection antigen, BALB/c female spleen cells were pulsed with 9m, 9wt, or an unrelated Kd-binding peptide derived from human c-erbB2. BALB/c female mice were immunized with 2 × 107 spleen cells, either peptide pulsed or nonpulsed, two times at 1-week intervals. Seven days after the second immunization, mice were challenged with 1 × 106 CMS5a cells injected subcutaneously. To facilitate immunization with peptides (32), 10 ng of IL-12 was injected intraperitoneally 13 times every other day from the day of first immunization. As shown in Fig. 4, mice immunized with syngeneic spleen cells pulsed with peptide 9m developed resistance to CMS5a challenge, whereas no tumor immunity was obtained in any of the other groups. IL-12 treatment was essential to show antitumor immunity in this system, because mice vaccinated with 9m-pulsed spleen cells in the absence of exogenous IL-12 showed no resistance to CMS5 challenge.
Figure 4.
Effect of vaccination with the mutated ERK2-derived peptide 9m on the growth of CMS5 cells. Spleen cells pulsed with 9m, 9wt, or a control erbB2-derived peptide, or nonpulsed naive spleen cells were injected subcutaneously in BALB/c female mice, five mice per group, on day 0 and day 7. Mice were challenged with 1 × 106 CMS5a cells subcutaneously on day 14, and tumor growth was measured. From day 0 to day 25, 10 ng of IL-12 was injected intraperitoneally every other day.
Conclusion.
Of the limited number of CTL-recognized peptide antigens defined in mouse tumor systems (33–39), the antigen encoded by the mutated ERK2 gene most closely resembles the characteristics one might expect for the individually distinct tumor rejection antigens of 3-MC-induced sarcomas. It has been suggested that the individually distinct tum− antigens of in vitro mutagenized tumor cells (40, 41) are counterparts of the diverse transplantation antigens of 3-MC-induced sarcomas. However, antigen-loss variants of tum− tumors appear to be far easier to isolate than antigen-loss variants of chemically induced sarcomas. In fact, the tumor rejection antigens of chemically induced tumors are quite stable despite prolonged transplantation in mice with a normal immune system. We have attempted without success to develop antigen-loss variants of CMS5 by forced passage in immunized mice and by repeated exposure in vitro to C18. This antigenic stability even in the face of strong immunoselection suggests that the tumor-rejection antigens of chemically induced tumors play an essential role in the maintenance of the malignant phenotype. Initial efforts to show a transforming activity of the mutated ERK2 gene have been unsuccessful. However, as CMS5 also has a mutation in p53, with an arginine-to-proline substitution at position 155, we are now investigating whether cotransfection of the mutated ERK2 gene with the CMS5 mutated p53 gene will result in transformation.
ABBREVIATIONS
- 3-MC
3-methylcholanthrene
- CTL
cytotoxic T lymphocytes
- TNF
tumor necrosis factor
- IL-12
interleukin 12
References
- 1.Gross L. Cancer Res. 1943;3:326–333. [Google Scholar]
- 2.Foley E J. Cancer Res. 1953;13:835–837. [PubMed] [Google Scholar]
- 3.Prehn R T, Main J M. J Natl Cancer Inst. 1957;18:769–778. [PubMed] [Google Scholar]
- 4.Klein G, Sjögren H O, Klein E, Hellström K E. Cancer Res. 1960;20:1561–1572. [PubMed] [Google Scholar]
- 5.Old L J, Boyse E A, Clarke D A, Carswell E A. Ann NY Acad Sci. 1962;101:80–106. [Google Scholar]
- 6.Globerson A, Feldman M. J Natl Cancer Inst. 1963;32:1229–1243. doi: 10.1093/jnci/32.6.1229. [DOI] [PubMed] [Google Scholar]
- 7.Basombrio M A, Prehn R T. Natl Cancer Inst Monogr. 1972;35:117–124. [PubMed] [Google Scholar]
- 8.Srivastava P K, Old L J. Immunol Today. 1988;9:78–83. doi: 10.1016/0167-5699(88)91269-8. [DOI] [PubMed] [Google Scholar]
- 9.Old L J, Boyse E A. Annu Rev Med. 1964;15:167–186. doi: 10.1146/annurev.me.15.020164.001123. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg P D. Adv Immunol. 1991;49:281–355. doi: 10.1016/s0065-2776(08)60778-6. [DOI] [PubMed] [Google Scholar]
- 11.Shiku H, Takahashi T, Bean M A, Old L J, Oettgen H F. J Exp Med. 1976;144:1116–1120. doi: 10.1084/jem.144.4.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duley M E, Roopenian D C. J Exp Med. 1996;184:441–447. doi: 10.1084/jem.184.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Old L J, Stockert E. Annu Rev Genet. 1977;11:127–160. doi: 10.1146/annurev.ge.11.120177.001015. [DOI] [PubMed] [Google Scholar]
- 14.DeLeo A B, Shiku H, Takahashi T, John M, Old L J. J Exp Med. 1977;146:720–734. doi: 10.1084/jem.146.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama E, Uenaka A, Stockert E, Obata Y. Cancer Res. 1984;44:5138–5144. [PubMed] [Google Scholar]
- 16.Lattime E C, Stoppacciaro A, Stutman O. J Immunol. 1988;141:3422–3428. [PubMed] [Google Scholar]
- 17.Van Pel A, De Plaen E, Boon T. Somatic Cell Genet. 1985;11:467–475. doi: 10.1007/BF01534840. [DOI] [PubMed] [Google Scholar]
- 18.Leo O, Foo M, Sachs D H, Samelson L E, Bluestone J A. Proc Natl Acad Sci USA. 1987;84:1374–1378. doi: 10.1073/pnas.84.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dialynas D P, Quan Z S, Wall K A, Pierres A, Quintans J, Loken M R, Pierres M, Fitch F W. J Immunol. 1983;131:2445–2451. [PubMed] [Google Scholar]
- 20.Nakayama E, Uenaka A. J Exp Med. 1985;161:345–355. doi: 10.1084/jem.161.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozato K, Mayer N M, Sachs D H. Transplantation. 1982;34:113–120. doi: 10.1097/00007890-198209000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Ozato K, Hansen T H, Sachs D H. J Immunol. 1980;125:2473–2477. [PubMed] [Google Scholar]
- 23.Ozato K, Takahashi H, Appella E, Sears D W, Murre C, Seidman J G, Kimura S, Tada N. J Immunol. 1985;134:1749–1758. [PubMed] [Google Scholar]
- 24.Kobayashi M, Fitz L, Ryan M, Hewick R M, Clark S C, Chan S, Loudon R, Sherman F, Perussia B, Trinchieri G. J Exp Med. 1989;170:827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gately M K, Warrier R R, Honasoge S, Carvajal D M, Faherty D A, Connaughton S E, Anderson T D, Sarmiento U, Hubbard B R, Murphy M. Int Immunol. 1994;6:157–167. doi: 10.1093/intimm/6.1.157. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama E, Shiku H, Takahashi T, Oettgen H F, Old L J. Proc Natl Acad Sci USA. 1979;76:3486–3490. doi: 10.1073/pnas.76.7.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traversari C, van der Bruggen P, Van den Eynde B, Hainaut P, Lemoine C, Ohta N, Old L J, Boon T. Immunogenetics. 1992;35:145–152. doi: 10.1007/BF00185107. [DOI] [PubMed] [Google Scholar]
- 28.Brichard V, Van Pel A, Wölfel T, Wölfel C, De Plaen E, Lethé B, Coulie P, Boon T. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dailey L, Basilico C. J Virol. 1985;54:739–749. doi: 10.1128/jvi.54.3.739-749.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seed B, Aruffo A. Proc Natl Acad Sci USA. 1987;84:3365–3369. doi: 10.1073/pnas.84.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Her J-H, Wu J, Rail T B, Sturgill T W, Weber M J. Nucleic Acids Res. 1991;19:3743. doi: 10.1093/nar/19.13.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noguchi Y, Richards E C, Chen Y T, Old L J. Proc Natl Acad Sci USA. 1995;92:2219–2223. doi: 10.1073/pnas.92.6.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lurquin C, Van Pel A, Mariamé B, De Plaen E, Szikora J-P, Janssens C, Reddehase M J, Lejeune J, Boon T. Cell. 1989;58:293–303. doi: 10.1016/0092-8674(89)90844-1. [DOI] [PubMed] [Google Scholar]
- 34.Lethé B, Van den Eynde B, Van Pel A, Corradin G, Boon T. Eur J Immunol. 1992;22:2283–2288. doi: 10.1002/eji.1830220916. [DOI] [PubMed] [Google Scholar]
- 35.Mandelboim O, Berke G, Fridkin M, Feldman M, Eisenstein M, Eisenbach L. Nature (London) 1994;369:67–71. doi: 10.1038/369067a0. [DOI] [PubMed] [Google Scholar]
- 36.Noguchi Y, Chen Y T, Old L J. Proc Natl Acad Sci USA. 1994;91:3171–3175. doi: 10.1073/pnas.91.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uenaka A, Ono T, Akisawa T, Wada H, Yasuda T, Nakayama E. J Exp Med. 1994;180:1599–1607. doi: 10.1084/jem.180.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang A Y C, Gulden P H, Woods A S, Thomas M C, Tong C D, Wang W, Engelhard V H, Pasternack G, Cotter R, Hunt D, Pardoll D M, Jaffee E M. Proc Natl Acad Sci USA. 1996;93:9730–9735. doi: 10.1073/pnas.93.18.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bloom M B, Perry-Lalley D, Robbins P F, Li Y, El-Gamil M, Rosenberg S A, Yang J C. J Exp Med. 1997;185:453–459. doi: 10.1084/jem.185.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boon T. Adv Cancer Res. 1983;39:121–151. doi: 10.1016/s0065-230x(08)61034-9. [DOI] [PubMed] [Google Scholar]
- 41.Van Pel A, Warnier G, Van den Eynde B, Lethé B, Lurquin C, Boon T. Ann NY Acad Sci. 1991;636:43–51. doi: 10.1111/j.1749-6632.1991.tb33437.x. [DOI] [PubMed] [Google Scholar]