Abstract
This study was initiated on the hypothesis that aryl acetic acid and aryl carboxylic acid-containing drugs would inhibit human phenol sulfotransferase (SULT1A1), and that isoform selectivity would depend on the interaction of the aryl portion of the molecule with the acceptor binding site of the sulfotransferase. This hypothesis was based on results with the rat orthologue enzyme showing that oxidation of phenolic substrates to carboxylic acid derivatives resulted in competitive inhibition of rat phenol sulfotransferase. We chose nine structurally representative non-steroidal anti-inflammatory agents and determined their inhibitory potency and selectivity toward human liver phenol sulfotransferase (SULT1A1) and expressed human estrogen sulfotransferase (SULT1E1). The results show that the tested agents reversibly inhibit human liver cytosolic phenol sulfotransferase activity with 50% inhibitory concentrations (IC50) ranging from 0.1 μM to 3800 μM. These agents also inhibited SULT1E1 (IC50 = 6 μM to 9000 μM). The agents were clearly isoform selective, with IC50 ratios (1E1/1A1) ranging from 0.01 to 200. Nimesulide, meclofenamate, and piroxicam were more selective towards SULT1A1 inhibition, while sulindac and ibuprofen were more selective towards SULT1E1 inhibition. Two agents lacking a carboxylic acid functional group, nimesulide and piroxicam, showed that the carboxylate could be substituted by enolate or methylsulfonamide and retain sulfotransferase inhibitory characteristics. Kinetic studies determined the type of inhibition of SULT1A1 for three agents (meclofenamate, nimesulide, aspirin) to be non-competitive or partial non-competitive versus both substrate (p-nitrophenol) and cofactor (PAPS). This inhibition mechanism indicates that meclofenamate, nimesulide and aspirin bind near enough to the substrate binding site to prevent catalysis but not affect dissociation of the substrate-enzyme complex. The inhibition of SULT1A1 by meclofenamate, nimesulide, salicylic acid and aspirin may be clinically relevant based on ratio of inhibition constant to predicted in vivo inhibitor concentration ([I]/IC50 >1).
Keywords: Human SULT1A1, human SULT1E1, human liver cytosol, non-steroidal anti-inflammatory, inhibition of sulfonation, sulfotransferase
INTRODUCTION
Phenol and estrogen sulfotransferase are members of the cytosolic sulfotransferases which preferentially catalyze sulfonation of phenolic functional groups on endogenous and xenobiotic compounds. Estrogen sulfotransferase (SULT1E1) has high affinity (Km = ∼20 nM) for endogenous steroids such as estradiol and estrone, and sulfonation of these estrogens is important for attenuation of steroid hormone signaling in tissues such as endometrium, mammary, and testes [1]. Phenol sulfotransferase, of which SULT1A1 is the major human member, is most well-known for conjugation of a diversity of small molecule phenols, hydroxylamines, and N-oxides. SULT1A1 converts many procarcinogens to reactive metabolites which form covalent adducts with DNA and are mutagenic in test systems [2]. SULT1A1 also aids clearance of many therapeutic drugs and environmental compounds and, conversely, catalyzes conversion of the prodrug minoxidil to the therapeutically active minoxidil-sulfate form. Inhibition of the cytosolic sulfotransferases would disrupt their normal action and mediate beneficial or adverse responses depending on the substrate and isoform involved.
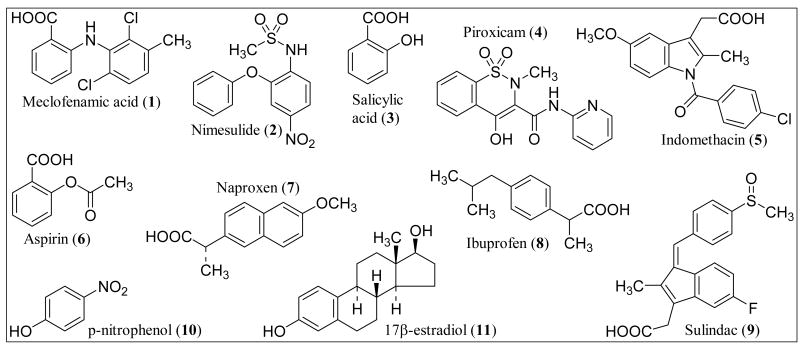
This study was initiated to help define the structural features necessary for inhibition of two major human cytosolic sulfotransferases, SULT1A1 and SULT1E1. We hypothesized that aryl acetic acid and aryl carboxylic acid-containing drugs would inhibit human phenol sulfotransferase (SULT1A1), and that isoform selectivity would depend on the interaction of the aryl portion of the molecule with the acceptor binding site of the sulfotransferase. This hypothesis was based on results with the rat orthologue enzyme showing that oxidation of phenolic substrates to carboxylic acid derivatives resulted in competitive inhibition of rat phenol sulfotransferase [3]. We selected nine compounds, shown in Fig. 1, from a large class of carboxylic acid-containing drugs, the non-steroidal anti-inflammatory agents. These agents all inhibit cyclooxygenase (COX-1, COX-2) with varying selectivity and potency. These nine compounds were selected for a combination of reasons. Naproxen and salicylic acid had been previously reported to inhibit human or rat sulfotransferases [3, 4]. Nimesulide and piroxicam are structurally distinct, replacing the traditional carboxylic acid functional group with a sulfonamide and enolate, respectively. Aspirin, ibuprofen, indomethacin, sulindac, piroxicam, and meclofenamate have been reported to exhibit cancer chemopreventive activity [5]. Due to the involvement of sulfotransferases in the bioactivation of the food-borne heterocyclic aromatic amine carcinogens [6] and other mutagens [2], sulfotransferase inhibition has been proposed as a mechanism of chemoprevention [7, 8]. While there is no direct evidence that sulfotransferase inhibition is involved in the chemoprevention reported by these agents, we thought it beneficial to include these reported chemopreventive agents in our study of aryl acetic acid and aryl carboxylic acid-containing drugs.
Fig. (1).
Structures of the nine non-steroidal anti-inflammatory agents used in this study (1-9). Compounds are numbered as to order of inhibition affinity toward SULT1A1. Compounds 10 and 11 are structures of the sulfotransferase substrates used in this study.
In summary, we aimed to determine whether representative aryl acetic acid and aryl carboxylic-acid containing drugs would inhibit human SULT1A1 or SULT1E1 in a selective manner. We used human liver cytosol as a source for SULT1A1 and cytosol from overexpressed E. coli XL-1 Blue transfected with pKK233-2-hEST as a source for SULT1E1. The results of our study demonstrate that structurally distinct non-steroidal anti-inflammatory agents inhibit human phenol and estrogen sulfotransferase with clear isoform-selectivity. This work represents the first report of human SULT1E1 inhibition by selected non-steroidal anti-inflammatory agents and is the first report of Ki determination for aspirin, meclofenamate or nimesulide against any sulfotransferase.
EXPERIMENTAL METHODS
Chemicals and Reagents
Ibuprofen, naproxen, salicylic acid, meclofenamate, sulindac, piroxicam, indomethacin, aspirin, nimesulide (all NSAIDs were USP grade), p-nitrophenol (spectrophotometric grade), and p-nitrophenylsulfate were purchased from Sigma Chemical Co. (St. Louis, MO).
3′-Phosphoadenosine-5′-phosphosulfate (PAPS) of the highest available grade (∼90%) was obtained from Sigma Chemical Co. and was used without further purification. Concentrated PAPS solutions (∼10 mM) were prepared and stored at -80°C until dilution just before use. As a control, we also purified the commercial PAPS preparation by ion exchange chromatography on DEAE-cellulose [9] and found no benefit for the sulfotransferase activity assays.
Radiolabeled [6,7-3H(N)]-estradiol (41.8 Ci/mmol) and [6,7-3H(N)]-estrone sulfate (57.3 Ci/mmol) were purchased from Perkin Elmer Life Sciences (Boston, MA) and stored in ethanol solution at -20°C. Estrone-sulfate was used in place of estradiol-sulfate to validate the separation method.
Transfected E. Coli XL-1 Blue cells and anti-SULT1E1 antibody were kindly provided by Dr. Charles N. Falany [10]. For Luria Broth preparation, 10 g sodium chloride, 10 g tryptone and 5 g yeast extract were dissolved in 1 L of deionized water. The pH was adjusted to 7.0 with 1 N sodium hydroxide and the medium autoclaved before use.
Enzyme preparations
Human liver cytosol
Cytosols were prepared as previously described [6] from human liver tissue obtained from the International Institute for the Advancement of Medicine with IRB approval. Cytosols were stored at -70°C in 50 mM Tris-HCl pH 7.8, 0.25 M sucrose, 0.5 mM EDTA, 0.1 mM DTT, and 0.02 nM BHT with no loss of activity from storage. Protein concentration of liver cytosol was determined by Biuret assay (Sigma Total Protein).
Expressed human estrogen sulfotransferase
Expressed human estrogen sulfotransferase was prepared from E. coli XL1-Blue cells transfected with bacterial expression vector pKK233-2-hEST [10] as supplied by Dr. CN Falany (Birmingham, AL). Suspension cultures in Luria Broth were inoculated from single colonies using standard microbiological techniques. After growing cells to late log phase (OD = 0.5-0.7) in the presence of ampicillin, the pKK233-2 vector was activated by addition of the promoter isopropyl-β-d-thiogalactopyranoside (IPTG, 0.3 mM). Optimal induction time for SULT1E1 expression was found to be 2 hours. Cell pellets harvested from 100 ml culture medium were lysed by sonication in 2 ml bacterial lysis buffer (50 mM Tris-HCl pH 7.4, 0.25 M sucrose, 1 mM EDTA, 10 mM DTT, 1 mM PMSF, 0.3 mg/ml lysozyme). After centrifugation at 100,000 × g for 1 hour at 4°C, the supernatant (cell cytosol) from the overexpressed bacterial culture was used as expressed human estrogen sulfotransferase for Bradford protein assay, SULT1E1 activity assay, and SDS-PAGE with Western Blot analysis. SULT1E1 protein expression was confirmed by SDS-PAGE with coomassie blue and Western Blot analysis using SULT1E1 antibody generously supplied by Dr. Falany [10].
Sulfotransferase assays
SULT1A1 activity
p-Nitrophenol sulfonation was determined with human liver cytosol as the enzyme source under conditions favoring selective catalysis by SULT1A1. Assay conditions were optimized to ensure initial rate conditions including linearity with protein concentration, linearity with incubation time, and excess of substrate and co-factor. Except as noted below, these conditions included 20 min incubation at 37°C in a total volume of 0.25 ml containing 4 μM p-nitrophenol, 20 μM PAPS, 2 mM potassium phosphate pH 7.4, and 0.16 mg/ml cytosolic protein. Reactions were started by addition of enzyme and stopped by addition of 0.025 ml of 4 M perchloric acid with centrifugation of the protein pellet. Formation of p- nitrophenylsulfate (pNPS) in the supernatant was determined after HPLC separation on a Hitachi D7000 system with isocratic elution (1 ml/min, 15% acetonitrile (v/v), 85% 0.05 M ammonium acetate pH 4.0 (v/v)) from a Waters Symmetry C18 column (5 μm, 3 × 150 mm) with detection at 280 nm [7]. pNPS concentrations were determined from a standard curve prepared from injection of known concentrations of pNPS (7 different concentrations bracketing the area of interest). Control experiments, in the absence of PAPS or in the absence of enzyme, showed no pNPS formation.
SULT1E1 activity
17β-estradiol sulfonation was determined with expressed human estrogen sulfotransferase as the enzyme source. Assay conditions were optimized to ensure initial rate conditions including linearity with protein concentration, linearity with incubation time, and excess of substrate and co-factor. Except as noted below, these conditions included 20 min incubation at 37°C in a total volume of 0.16 ml containing 20 nM estradiol ([6,7-3H(N)], 41.8 Ci/mmol), 0.4 μM PAPS, 11 mM potassium phosphate pH 6.5, 1.56 mM magnesium chloride, 14 mM DTT, 1 mg/ml BSA, and 0.14 μg/ml expressed enzyme cytosol. Reactions were started by addition of enzyme and stopped by addition of 0.5 ml dichloromethane. Formation of estradiol-sulfate was determined after extraction of estradiol into the organic phase. After vigorous vortex mixing and centrifugation (600 × g) to separate the phases, an aliquot (40 μl) of the aqueous phase was mixed with 0.2 ml SuperMix cocktail (PerkinElmer) in a 24-well PET sample plate (Wallac) and taken for liquid scintillation counting on a Wallac 1450 MicroBeta TriLux counter. Control experiments were conducted to ensure that radiolabeled estradiol was completely extracted from the aqueous phase (incubation in absence of enzyme) and that minimal radiolabeled estradiol-sulfate was lost into the organic phase.
Inhibition assays
For assays determining 50% inhibitory constants (IC50) for SULT1A1, p-nitrophenol sulfonation was measured as described above in the presence of a wide range of concentrations of each potential inhibitor (0.02 μM – 1 mM), as appropriate. For assays determining IC50 for SULT1E1, 17β-estradiol sulfonation was measured as described above in the presence of a wide range of concentrations of each potential inhibitor (5 nM – 5 mM), as appropriate. Salicylic acid, ibuprofen, naproxen, and meclofenamate were dissolved in 95% ethanol to a concentration of 125 mM. Piroxicam, indomethacin, and nimesulide were dissolved (125 mM) in dimethylsulfoxide. After dilution into the incubation mixture, final solvent concentrations were 0.8% (v/v) or lower and did not affect enzyme activity. Aspirin was dissolved in incubation buffer.
For assays determining kinetic inhibition constants (Ki, Km, Vmax) for SULT1A1, p-nitrophenol sulfonation was measured while varying substrate (pNP), cofactor (PAPS), and inhibitor concentrations. When pNP was varied (0.5, 1, 2, 4 μM), the concentration of PAPS was held constant at 20 μM. When PAPS was varied (0.25, 0.3, 0.45, 0.7, 1, 1.6, 5 μM), the concentration of pNP was held constant at 4 μM. Incubation time for all kinetic assays was reduced to 10 min to provide initial rate conditions at the lowest substrate/cofactor concentrations. All other conditions remained as described for SULT1A1 activity. The inhibitor concentrations were determined empirically to produce approximately 75% and 50% of maximal enzyme activity in absence of inhibitor: 0.02 μM and 0.04 μM for meclofenamate, 0.3 μM and 0.6 μM for nimesulide, 1 μM and 540 μM for aspirin.
To confirm that the inhibition produced by these non-steroidal anti-inflammatory agents was reversible, we pre-incubated the enzyme (4 mg/ml in incubation buffer) for 10 min at 37°C in the presence of (a) no additions, (b) inhibitor, (c) inhibitor plus 20 μM PAPS, (d) inhibitor plus 4 μM pNP, and (e) inhibitor plus 20 μM PAPS plus 4 μM pNP. Inhibitors were at concentrations near their calculated Ki. After each pre-incubation, an aliquot was withdrawn and used immediately as the source of enzyme in a standard assay for phenol sulfotransferase activity [11]. After 40-fold dilution into the standard assay, protein concentration (0.1 mg/ml) was similar to that used for the activity assays described above. When the inhibition is truly reversible, enzyme activity does not change upon pre-incubation of inhibitor with enzyme (conditions b-e). This approach is similar to an assay we used previously to detect irreversible inhibition of rat liver phenol sulfotransferase [11].
Data analysis
IC50 calculations
All assays were performed in triplicate and data is reported as mean and standard deviation of these three determinations. Product formation data was converted to % activity (100% = activity in absence of inhibitor) and plotted versus log of inhibitor concentration. The resulting data values were fit by non-linear regression to the equation (y = a / (1 + exp(-(x-x0)/b))) for a sigmoidal curve, and the inhibitor concentrations corresponding to 50% activity were determined from the regression parameters. Goodness of fit was evaluated using the standard deviation of the residuals and R2 for each dataset. The data for SULT1A1 inhibition by meclofenamate and nimesulide did not fit a sigmoidal curve in the region of inhibitor concentrations tested, and a logarithmic function was used instead (y = y0 + a * ln(x)) for regression.
Calculations for Kinetic constants
All assays were performed in triplicate and data is reported as mean and standard deviation of these three determinations. Kinetic data was evaluated using a non-linear curve fitting program specializing in enzyme kinetics (Enzyme Kinetics module, SigmaPlot 8.0). The data was fit by non-linear regression to the eight equations modeling reversible inhibition (competitive, non-competitive, uncompetitive, and mixed (full and partial for each). The best fit of the data was determined by evaluation of the Akaike Criterion for each equation. We repeated each kinetic experiment at least four times resulting in similar calculated kinetic constants and best-fit equation.
RESULTS
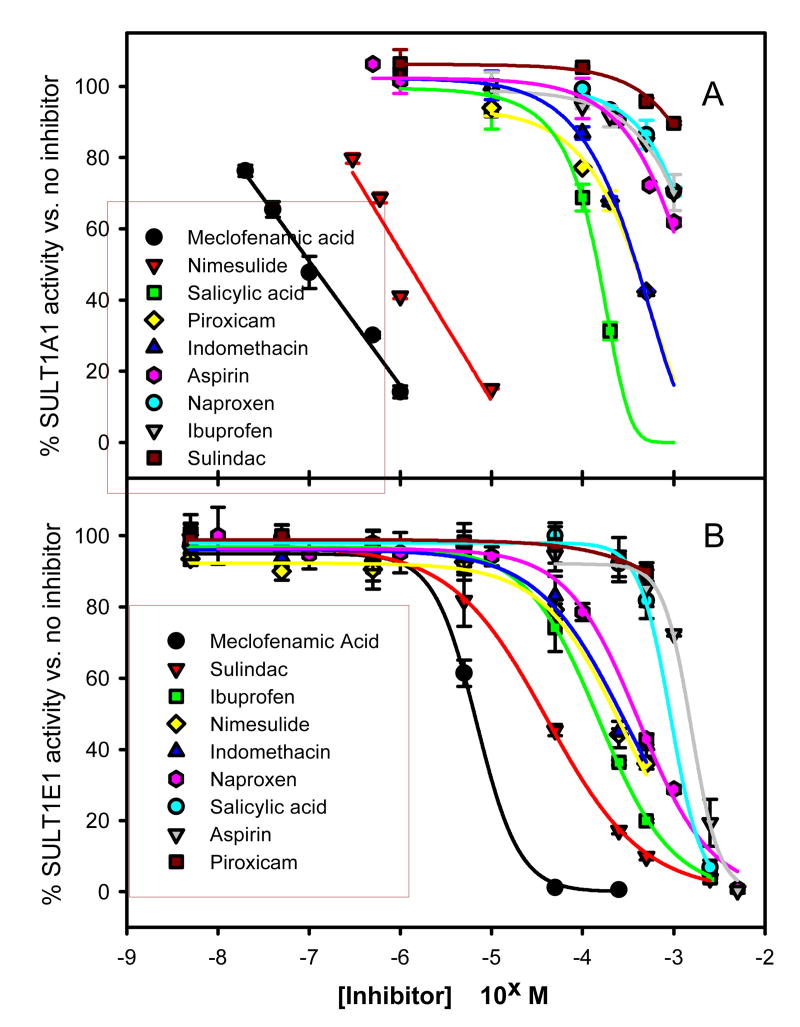
Nine representative carboxylic acid-containing drugs were analyzed for their ability to inhibit human liver phenol sulfotransferase (SULT1A1 activity) and expressed human estrogen sulfotransferase (SULT1E1 activity). Structures of these agents along with the sulfotransferase substrates are shown in Fig. 1. We measured enzyme activity in the presence of a wide range of concentrations of each potential inhibitor (1 nM – 5 mM), as appropriate, and calculated 50% inhibitory concentration (IC50) using a sigmoidal regression curve as described in the methods. We found that each of the nine compounds inhibited both sulfotransferase isoforms with clear isoform selectivity (Fig. 2, Table 1). Nimesulide, meclofenamate and piroxicam were more selective toward SULT1A1 inhibition. Sulindac and ibuprofen were more selective toward SULT1E1 inhibition. Meclofenamate was the most potent inhibitor of both SULT1A1 and SULT1E1; however, the difference in inhibition potency made it selective toward SULT1A1 (SULT1E1/SULT1A1 IC50 ratio = 60). Nimesulide was second most potent inhibitor of SULT1A1 and was most selective toward SULT1A1 inhibition (IC50 ratio = 190). Even though piroxicam had intermediate inhibition potency toward SULT1A1 (IC50 = 380 μM), it was a very weak inhibitor of SULT1E1 resulting in selectivity toward SULT1A1 (IC50 ratio = 25). After meclofenamate, the most potent inhibitors of SULT1E1 were sulindac and ibuprofen, and these were selective toward SULT1E1 (IC50 ratios 0.01, 0.07). Salicylic acid, indomethacin, aspirin and naproxen inhibited both isoforms with less than 10-fold selectivity (Table 1) and moderate potency (IC50 = 150 – 1700 μM). Two agents lacking a carboxylic acid functional group, nimesulide and piroxicam, showed that the carboxylate could be substituted by enolate or methylsulfonamide and retain sulfotransferase inhibitory characteristics.
Fig. (2).
Inhibition of SULT1A1 or SULT1E1 activity by representative non-steroidal anti-inflammatory agents. Symbols represent data and line represents non-linear regression fit as described in methods section. Calculated IC50 values are summarized in Table 1.
Table 1. Inhibition of estrogen (SULT1E1) or phenol (SULT1A1) sulfotransferase activity by selected non-steroidal anti-inflammatory agents.
Inhibitor concentrations resulting in 50% reduction of control sulfotransferase activity (IC50) were calculated by non-linear regression as described in methods. Data and regression curves shown in Fig. (2).
| Inhibitor (structure #) | SULT 1A11 IC50 μM (vs. 4 μM pNP) |
SULT 1E1
IC50 μM (vs. 20 nM E2) |
IC50 Ratio
1E1/1A1 |
|---|---|---|---|
| Meclofenamic Acid (1) | 0.11 | 6.5 | 60 |
| Nimesulide (2) | 1.2 | 230 | 190 |
| Salicylic Acid (3) | 150 | 910 | 6 |
| Piroxicam (4) | 380 | 94002 | 25 |
| Indomethacin (5) | 390 | 270 | 0.7 |
| Aspirin (6) | 1300 | 1500 | 1.1 |
| Naproxen (7) | 1700 | 190 | 0.1 |
| Ibuprofen (8) | 1900 | 140 | 0.07 |
| Sulindac (9) | 38002 | 38 | 0.01 |
IC50 values for SULT1A1 represent a single human liver cytosol; this same sample was used to obtain kinetic data in Table 2. Multiple human liver cytosols were used for IC50 determinations with similar calculated values for each (within 10%).
The IC50 values reported for piroxicam (SULT1E1) and sulindac (SULT1A1) are estimates based on non-linear regression of maximal inhibition of 10% (see Fig. 2).
To assess whether these compounds were likely to cause sulfotransferase inhibition in vivo at clinically relevant doses, it is generally accepted that the best prediction method is the [I]/Ki ratio [reviewed in 12], where [I] is the estimated in vivo concentration of the inhibitor. By this prediction method, inhibitors with high risk of causing clinically significant inhibition in vivo will have [I]/Ki > 1.0; inhibitors with low risk will have [I]/Ki < 0.1; and inhibitors of medium risk will have intermediate [I]/Ki. One challenge of using this prediction method is that several options exist for estimating [I]. For our estimations, we used published steady-state peak plasma concentration, Cmax, for [I] (Table 2, [13]). A second challenge of using [I]/Ki ratio for prediction of clinical relevancy of inhibition is that one often prefers to make this prediction using the experimentally more accessible IC50 constants, before conducting the extensive kinetic experiments necessary for direct Ki determination. Ki can be calculated from the IC50 based on mathematical relationship. However, the proper equation depends upon knowing the mechanism of inhibition [12], and the same extensive kinetic experiments needed for Ki determination are needed for mechanism determination. Thus, before Ki determination on a subset of compounds, we assessed the likelihood of these inhibitors to inhibit sulfotransferase activity in vivo by direct comparison of measured IC50 with predicted [I] (published Cmax) (Tables 1-2). Because the kinetic constants were determined in the presence of excess protein (human liver cytosol or BSA added to expressed protein), the Cmax values were not corrected for plasma protein binding. This direct comparison predicts that meclofenamate, nimesulide, salicylic acid and aspirin may significantly inhibit human SULT1A1 activity in vivo at typical therapeutic doses ([I]/IC50 >1). The direct comparison of SULT1E1 IC50 and [I] predicts that meclofenamate, nimesulide, salicylic acid, aspirin, naproxen and ibuprofen may cause mild in vivo inhibition of human SULT1E1 at typical therapeutic doses ([I]/IC50∼ 1).
Table 2. Parameters used for prediction of likelihood for in vivo inhibition of sulfotransferase activity at clinically relevant doses of each NSAID.
Calculations and choice of parameters discussed in Results section.
| Inhibitor (structure #) | [I]
Cmax (μM) 1 |
SULT1A1
[I] / IC502 |
SULT1A1
[I] / Ki3 |
SULT1E1
[I] / IC502 |
|---|---|---|---|---|
| Meclofenamate (1) | 16 | 140 | 320 | 2.5 |
| Nimesulide (2) | 220 | 180 | 310 | 1 |
| Salicylic Acid (3) | 2100 4 | 14 | 2 | |
| Piroxicam (4) | 35 | 0.09 | < 0.004 | |
| Indomethacin (5) | 6 | 0.02 | 0.02 | |
| Aspirin (6) | 1600 | 1.2 | 1143 | 1 |
| Naproxen (7) | 410 | 0.24 | 2 | |
| Ibuprofen (8) | 290 | 0.15 | 2 | |
| Sulindac (9) | 11 | < 0.003 | 0.3 |
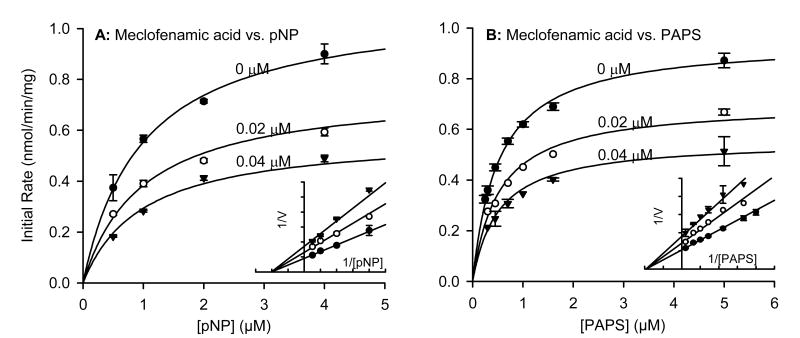
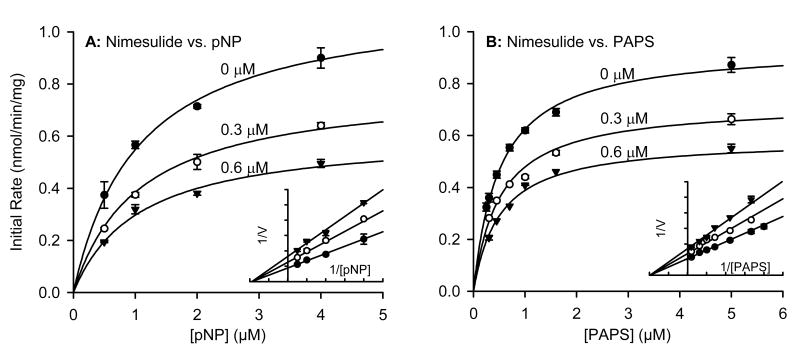
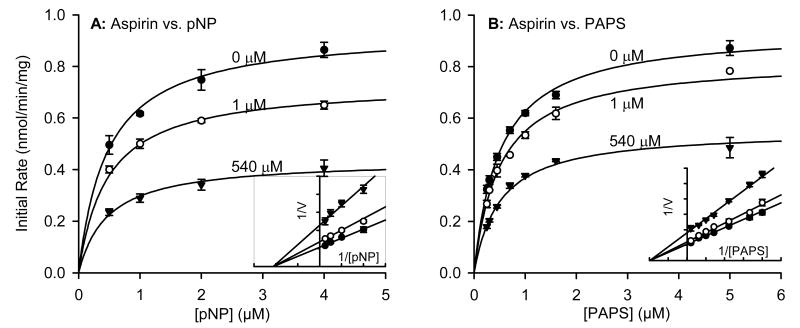
To determine the type of inhibition produced by these non-steroidal anti-inflammatory agents, we first confirmed that the inhibition was reversible as described in the methods. We then chose three compounds for detailed kinetic inhibition experiments with SULT1A1: aspirin, nimesulide and meclofenamate. These three, plus salicylic acid, were the only compounds tested for which the IC50 versus SULT1A1 was similar to or lower than the reported maximal plasma concentration (Cmax) after typical therapeutic dose (Table 2). We first varied acceptor substrate (pNP) concentration in the presence of increasing concentrations of each inhibitor and a constant saturating concentration of PAPS (20 μM) (Figs. 3A, 4A, 5A). After repeating the experiment several times to ensure reproducibility, we analyzed the data using a non-linear curve fitting program specializing in enzyme kinetics (Enzyme Kinetics module, SigmaPlot 8.0). The pNP concentrations were below those known to cause substrate inhibition, and the data followed apparent Michaelis-Menton kinetics. The apparent Km values for human liver cytosolic SULT1A1 activity (Table 3) in the absence of inhibitor were similar to those published for expressed SULT1A1*1 and SULT1A1*2 [14]. Non-linear regression showed that the inhibition data versus pNP as the variable substrate best fit the equation modeling non-competitive (nimesulide and meclofenamate) or partial non-competitive (aspirin) enzyme inhibition. Since none of the inhibitors proved to bind competitively with pNP, we also varied donor substrate (PAPS) in the presence of increasing concentrations of each inhibitor and a constant concentration of pNP (4 μM) (Figs. 3B, 4B, 5B). Again, non-linear regression showed that the inhibition data versus PAPS as the variable substrate best fit the equation modeling non-competitive (nimesulide and meclofenamate) or partial non-competitive (aspirin) enzyme inhibition. The calculated kinetic constants are summarized in Table 3.
Fig. (3).
Effect of meclofenamate (1) on kinetics of SULT1A1 activity in human liver cytosol. (A) Saturation curves versus varying concentrations of substrate p-nitrophenol. (B) Saturation curves versus varying concentrations of cofactor PAPS. Inserts show data plotted in Lineweaver-Burke format. Symbols represent data and line represents non-linear regression fit as described in methods section. Calculated kinetic parameters are summarized in Table 2.
Fig. (4).
Effect of nimesulide (2) on kinetics of SULT1A1 activity in human liver cytosol. (A) Saturation curves versus varying concentrations of substrate p-nitrophenol. (B) Saturation curves versus varying concentrations of cofactor PAPS. Inserts show data plotted in Lineweaver-Burke format. Symbols represent data and line represents non-linear regression fit as described in methods section. Calculated kinetic parameters are summarized in Table 2.
Fig. (5).
Effect of aspirin (6) on kinetics of SULT1A1 activity in human liver cytosol. (A) Saturation curves versus varying concentrations of substrate p-nitrophenol. (B) Saturation curves versus varying concentrations of cofactor PAPS. Inserts show data plotted in Lineweaver-Burke format. Symbols represent data and line represents non-linear regression fit as described in methods section. Calculated kinetic parameters are summarized in Table 2.
Table 3. Apparent kinetic constants for SULT1A1 activity.
Kinetic constants were calculated by nonlinear regression as described in methods. Data and regression curves shown in Figs. (3, 4, 5).
| Inhibitor (structure #) | pNP as variable substrate | PAPS as variable substrate | ||||
|---|---|---|---|---|---|---|
| Ki μM |
Km μM |
Vmax nmol(min*mg)-1 |
Ki μM |
Km μM |
Vmax nmol(min*mg)-1 |
|
| Meclofenamic acid (1) | 0.045 ± 0.003 | 0.99 ± 0.08 | 1.10 ± 0.03 | 0.056 ± 0.004 | 0.52 ± 0.03 | 0.95 ± 0.02 |
| Nimesulide (2) | 0.71 ± 0.05 | 1.10 ± 0.09 | 1.10 ± 0.04 | 0.99 ± 0.07 | 0.51 ± 0.03 | 0.94 ± 0.02 |
| Aspirin (6) | 1.4 ± 0.2 | 0.47 ± 0.05 | 0.94 ± 0.03 | 2.4 ± 0.5 | 0.50 ± 0.03 | 0.94 ± 0.02 |
By definition, non-competitive inhibitors bind close to the active site and change the shape of the active site such that the substrate can bind but the enzyme cannot convert bound substrate into product [15, 16]. Thus, Vmax is decreased in the presence of non-competitive inhibitor but the Km is not affected. This is typically interpreted to indicate that, while the non-competitive inhibitor does not affect the dissociation complex of the enzyme-substrate complex (Km), the substrate cannot be converted to product in the presence of inhibitor. The situation in partial non-competitive enzyme inhibition is similar with one important difference: the distance between the lines of the Lineweaver-Burke plot of a partial non-competitive inhibitor is not proportional with inhibitor concentration. This characteristic can be observed by comparing Figs. 3-4 with Fig. 5. This is typically interpreted to indicate that binding of the partial non-competitive enzyme inhibitor slows, but does not completely prevent, catalysis. Another characteristic of non-competitive inhibition is that the inhibition is not overcome in the presence of higher concentrations of substrate (Figs. 3-5).
DISCUSSION
We found that each of the nine representative non-steroidal anti-inflammatory agents inhibited both sulfotransferase isoforms with clear isoform selectivity (Fig. 2, Table 1). Nimesulide, meclofenamate and piroxicam were more selective toward SULT1A1 inhibition. Sulindac and ibuprofen were more selective toward SULT1E1 inhibition. Salicylic acid, indomethacin, aspirin, and naproxen inhibited both isoforms with less than 10-fold selectivity. While these compounds differ in volume, planarity, and shape, they all contain aromatic rings and are within the size range of known substrates of SULT1A1 and SULT1E1. Since it is difficult to visualize the interactions without a 3D model, we are currently modeling the binding of these and other inhibitors into binding sites developed from the X-ray crystal structures of SULT1A1 [17] and SULT1E1 [18] using Accelrys Insight II software. These modeling results are beyond the scope of this article.
More than half of all sulfotransferase inhibitors for which kinetic analyses have been published were determined to exhibit non-competitive or mixed (non-competitive and competitive) inhibition [reviewed in 19]. This is an unusually high fraction given that most of these inhibitors have structural similarities to sulfotransferase substrates and were expected to compete with acceptor substrate for binding to the enzyme. By definition, non-competitive inhibitors bind close to the active site and change the shape of the active site such that the substrate can bind but the enzyme cannot convert bound substrate to product [15, 16]. The propensity for non-competitive sulfotransferase inhibition seems more rational in light of studies showing that the human phenol sulfotransferase substrate binding site is large enough to bind two molecules of substrate simultaneously: an X-ray crystal structure of SULT1A1 [17] and a homology model of SULT1A3 [20] place two molecules of substrate bound into the substrate binding pocket. While it is not known whether the crystal or computational structure represents a kinetically active conformation, a substrate binding pocket large enough to accommodate two-substrates or one substrate and one non-competitive inhibitor could explain the propensity of sulfotransferases for both substrate inhibition and non-competitive inhibition. Kinetic data supporting two-substrate binding includes a kinetic study [21] which determined that two estradiol molecules bind per SULT1E1 monomer. This structural and kinetic evidence supports the conclusion that many, if not all, of the non-competitive sulfotransferase inhibitors bind to a sufficiently large substrate binding site such that both substrate and non-competitive inhibitor can be accommodated at the same time.
SULT1A1 exists as three major allozymes, SULT1A1*1, SULT1A1*2, SULT1A1*3. The *1 is considered wild-type and the variants differ by one amino acid each. The wild-type and *2 variant do not differ in Km for either p-nitrophenol or PAPS, indicating no difference in active site conformation [14]. The *3 variant has lower affinity for both p-nitrophenol and PAPS, as indicated by higher Km's [14]. The SULT1A1 genotype of our human liver cytosols are not known, but our detailed kinetic studies using a single human liver cytosol sample produced apparent Km values in agreement with those published for expressed SULT1A1*1 and SULT1A1*2 [14]. We also examined inhibition using cytosols from several different individuals, including both ‘high’ and ‘low’ activity samples, and found similar IC50 values indicating no difference in inhibitor binding amongst individual samples. It is possible, though we believe unlikely, that our kinetic measurements using human liver cytosol as enzyme source are detecting contributions to p-nitrophenol sulfonation from a second enzyme. Human liver contains two forms of phenol sulfotransferase, SULT1A1 and SULT1A2 [14]. However, at p-nitrophenol concentration of 4 μ M, it is not likely that SULT1A2 is contributing to our measured product formation because the Km of SULT1A2 for pNP is much higher than 4 μM [14].
After this study was initiated, Vietri et al [22,23] reported an overlapping set of non-steroidal anti-inflammatory agents inhibiting SULT1A1 activity with different IC50 values from those found in our study. Interestingly, the Km values for both pNP and PAPS in the current study are similar to those published for expressed SULT1A1*1 and SULT1A1*2 allozymes [14], while the Km values of Vietri et al [22] are similar to those reported previously for SULT1A1*3 [14]. Thus, different genotype of the enzyme source is a probable cause of the differences in calculated IC50 values for SULT1A1 inhibition between the two studies for the overlapping compounds (piroxicam, ibuprofen, naproxen, indomethacin, salicylic acid, nimesulide, meclofenamate).
In summary, each of the nine agents inhibited both human sulfotransferase isoforms, SULT1A1 and SULT1E1. Meclofenamate, nimesulide and piroxicam were more selective toward SULT1A1 inhibition, while sulindac and ibuprofen were more selective toward SULT1E1 inhibition. The type of inhibition of SULT1A1 was determined for three agents (meclofenamate, nimesulide, aspirin) to be non-competitive or partial non-competitive versus both substrate (pNP) and cofactor (PAPS). This inhibition mechanism indicates that meclofenamate, nimesulide and aspirin bind near enough to the substrate binding site to prevent catalysis but not affect dissociation of the substrate-enzyme complex. The inhibition of SULT1A1 by meclofenamate, nimesulide, salicylic acid and aspirin may be clinically relevant based on ratio of inhibition constant to predicted in vivo inhibitor concentration ([I]/IC50 >1). This is the first report of Ki determination for aspirin, meclofenamate or nimesulide against any sulfotransferase, and is the first report of human SULT1E1 inhibition by selected non-steroidal anti-inflammatory agents.
Acknowledgments
This research was supported by the following awards to RSK: The Rhode Island Foundation, Medical Research Grant #20020738; AACP Pharmacy Faculty New Investigator Award supported by The Burroughs Wellcome Fund and the American Foundation for Pharmaceutical Education; University of Rhode Island Council for Research; and NIH Grant Number P20 RR016457 from the BRIN/INBRE Program of the National Center for Research Resources. A portion of the work has been presented in abstract form: Drug Metab. Rev. 32(S2):192 (2000), Drug Metab. Rev. 33(S1):260 (2001).
Abbreviations
- BHT
butylated hydroxytoluene
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- IC50
50% inhibitory concentrations
- IPTG
isopropyl-β-D-thiogalactopyranoside
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- PMSF
phenylmethylsulfonyl fluoride
- pNP
p-nitrophenol
- SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- SULT1A1
human sulfotransferase isoform 1A1
- SULT1E1
human sulfotransferase isoform 1E1
Footnotes
The authors received no financial contributions for the work described and report no conflicts of interest.
References
- 1.Song WC. Biochemistry and reproductive endocrinology of estrogen sulfotransferase. Ann NY Acad Sci. 2001;948:43–50. doi: 10.1111/j.1749-6632.2001.tb03985.x. [DOI] [PubMed] [Google Scholar]
- 2.Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000;129:141–70. doi: 10.1016/s0009-2797(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 3.Rao SI, Duffel MW. Inhibition of aryl sulfotransferase by carboxylic acids. Drug Metab Dispos. 1991;19:543–545. [PubMed] [Google Scholar]
- 4.Harris RM, Hawker RJ, Langman MJS, Singh S, Waring RH. Inhibition of phenolsulfotransferase by salicylic acid: a possible mechanism by which aspirin may reduce carcinogenesis. Gut. 1998;42:272–275. doi: 10.1136/gut.42.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Agency for Research on Cancer (IARC) IARC Handbooks of Cancer Prevention. Lyon (France): IARC Press; 1997. Non-steroidal anti-inflammatory drugs. [Google Scholar]
- 6.King RS, Teitel CH, Kadlubar FF. In vitro bioactivation of N-hydroxy-2-amino-alpha-carboline. Carcinogenesis. 2000;21:1347–1354. [PubMed] [Google Scholar]
- 7.Eaton EA, Walle UK, Lewis AJ, Hudson T, Wilson AA, Walle T. Flavonoids, potent inhibitors of the human P-form phenolsulfotransferase. Potential role in drug metabolism and chemoprevention. Drug Metab Dispos. 1996;24:232–7. [PubMed] [Google Scholar]
- 8.Ghazali RA, Waring RH. The effects of flavonoids on human phenolsulphotransferases: Potential in drug metabolism and chemoprevention. Life Sci. 1999;65:1625–32. doi: 10.1016/s0024-3205(99)00423-3. [DOI] [PubMed] [Google Scholar]
- 9.King RS, Duffel MW. Oxidation-dependent inactivation of aryl sulfotransferase IV by primary N-hydroxy arylamines during in vitro assays. Carcinogenesis. 1997;18:843–849. doi: 10.1093/carcin/18.4.843. [DOI] [PubMed] [Google Scholar]
- 10.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Molec Biol. 1995;52:529–539. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- 11.Duffel MW, Modi RB, King RS. Interactions of a primary N-hydroxy arylamine with rat hepatic aryl sulfotransferase IV. Drug Metab Dispos. 1992;20:339–341. [PubMed] [Google Scholar]
- 12.Bachmann KA. Inhibition constants, inhibitor concentrations and the prediction of inhibitory drug drug interactions: pitfalls, progress, and promise. Current Drug Metabolism. 2006;7:1–14. doi: 10.2174/138920006774832541. [DOI] [PubMed] [Google Scholar]
- 13.Micromedex® Healthcare Series: DRUGDEX® System. Thomson Micromedex; Greenwood Village, Colorado: (Edition expires [date]). Pharmacokinetics: Drug concentration levels. [Google Scholar]
- 14.Raftogianis RB, Wood TC, Weinshilboum RM. Human phenol sulfotransferases SULT1A2 and SULT1A1: Genetic polymorphisms, allozymes properties, and human liver genotype-phenotype correlations. Biochem Pharmacol. 1999;58:605–616. doi: 10.1016/s0006-2952(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 15.Enzyme Kinetics help module, version 1.1. SPSS Science, Inc; Chicago, IL: 2001. SigmaPlot 2002, version 8.0. [Google Scholar]
- 16.Cleland WW. Steady state kinetics. In: Boyer PD, editor. The Enzymes. 3rd. Vol. 2. New York: Academic Press; 1970. pp. 18–21. [Google Scholar]
- 17.Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, McManus ME, Martin JL. Structure of a human carcinogen-converting enzyme, SULT1A1: Structural and kinetic implications of substrate inhibition. J Biol Chem. 2003;278:7655–7662. doi: 10.1074/jbc.M207246200. [DOI] [PubMed] [Google Scholar]
- 18.Shevtsov S, Petrotchenko EV, Pedersen LC, Negishi M. Crystallographic analysis of a hydroxylated polychlorinated biphenyl (OH-PCB) bound to the catalytic estrogen binding site of human estrogen sulfotransferase. EHP. 2003;111:884–888. doi: 10.1289/ehp.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang LQ, James MO. Inhibition of sulfotransferases by xenobiotics. Current Drug Metabolism. 2006;7:83–104. doi: 10.2174/138920006774832596. [DOI] [PubMed] [Google Scholar]
- 20.Barnett AC, Tsvetanov S, Gamage N, Martin JL, Duggleby RG, McManus ME. Active site mutations and substrate inhibition in human sulfotransferase 1A1 and 1A3. J Biol Chem. 2004;279:18799–18805. doi: 10.1074/jbc.M312253200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang HP, Varlamova O, Vargas FM, Falany CN, Leyh TS. Sulfuryl transfer: The catalytic mechanism of human estrogen sulfotransferase. J Biol Chem. 1998;273:10888–10892. doi: 10.1074/jbc.273.18.10888. [DOI] [PubMed] [Google Scholar]
- 22.Vietri M, De Santi C, Pietrabissa A, Mosca F, Pacifici GM. Inhibition of human liver phenol sulfotransferase by nonsteroidal anti-inflammatory drugs. Eur J Clin Pharmacol. 2000;56:81–87. doi: 10.1007/s002280050725. [DOI] [PubMed] [Google Scholar]
- 23.Vietri M, De Santi C, Pietrabissa A, Mosca F, Pacifici GM. Fenamates and the potent inhibition of human liver phenol sulphotransferase. Xenobiotica. 2000;30:111–116. doi: 10.1080/004982500237712. [DOI] [PubMed] [Google Scholar]