Summary
Ion pairs are ubiquitous in X-ray structures of coiled coils, and mutagenesis of charged residues can result in large stability losses. By contrast, pKa values determined by NMR in solution often predict only small contributions to stability from charge interactions. To help reconcile these results we used triple-resonance NMR to determine pKa values for all groups that ionize between pH 1 and 13 in the 33-residue leucine zipper fragment, GCN4p. In addition to the native state we also determined comprehensive pKa values for two models of the GCN4p denatured state: the protein in 6 M urea, and unfolded peptide fragments of the protein in water. Only residues that form ion pairs in multiple X-ray structures of GCN4p gave large pKa differences between the native and denatured states. Moreover, electrostatic contributions to stability were not equivalent for oppositely charged partners in ion pairs, suggesting that the interactions between a charge and its environment are as important as those within the ion pair. The pH dependence of protein stability calculated from NMR-derived pKa values agreed with the stability profile measured from equilibrium urea-unfolding experiments as a function of pH. The stability profile was also reproduced with structure-based continuum electrostatic calculations, although contributions to stability were overestimated at the extremes of pH. We consider potential sources of errors in the calculations, and how pKa predictions could be improved. Our results show that although hydrophobic packing and hydrogen bonding have dominant roles, electrostatic interactions also make significant contributions to the stability of the coiled coil.
Keywords: protein folding, ionization constant, salt-bridges, alpha-helix, finite difference Poisson-Boltzmann
Introduction
Electrostatic interactions play roles in protein folding, molecular recognition, and catalytic activity 1-4. A better understanding of these forces is important for developing improved potential energy functions for use in applications such as protein structure prediction and protein design 4-9. The pKa values of individual ionizable sites depend on Coulomb effects arising from interactions between charges, on charge-induced polarization effects, on interactions with other polar groups, and on interactions with water 2,4,6,10. These factors are currently difficult to evaluate experimentally in quantitative detail. Although steady progress is being made, the goal of predicting the magnitudes of electrostatic effects and their contributions to protein stability from protein structure remains to be fully realized. In the present work we have used NMR to obtain a comprehensive characterization of the ionization equilibria of the GCN4 leucine zipper in both its native (N) and denatured (D) states. The virtually complete set of pKa values for ionizable groups in the leucine zipper sheds new light on the contributions of electrostatic forces to structural stability, a subject that has generated considerable interest and debate 11-14. The pKa data additionally serves as a benchmark to test computational methods for structure-based calculation of electrostatic effects, and suggests possible sources of disparity between theory and experiment.
Coiled coils are oligomeric assemblies of two to five α-helices, with sequences organized in highly characteristic heptad repeats 15. The periodicity of α-helical structure supports intra-chain electrostatic interactions between residues with an i, i+3 and i, i+4 spacing 16. Packing of α-helix monomers in the coiled coil lends itself to additional inter-chain i, i+5’ electrostatic interactions 17,18. Inter-and intra-molecular hydrogen-bonded salt bridges are prevalent in the X-ray structure of the GCN4 leucine zipper (Fig. 1), and other coiled coils 12,15,19-21. By contrast NMR studies have shown that ionization of these residues often contributes little to the stability of the N state 17,22-24. On the surface of a protein, the Coulomb interaction energy favoring ion pair formation may be insufficient to balance out the entropic forces that favor breaking the salt bridge and the favorable hydration energy of the individual charges 1,25. Replacing charged residues with neutral analogues 11,13,26 often leads to large decreases in stability but the effects of the mutation may extend beyond the replacement of a charge 11-13,26.
Figure 1.
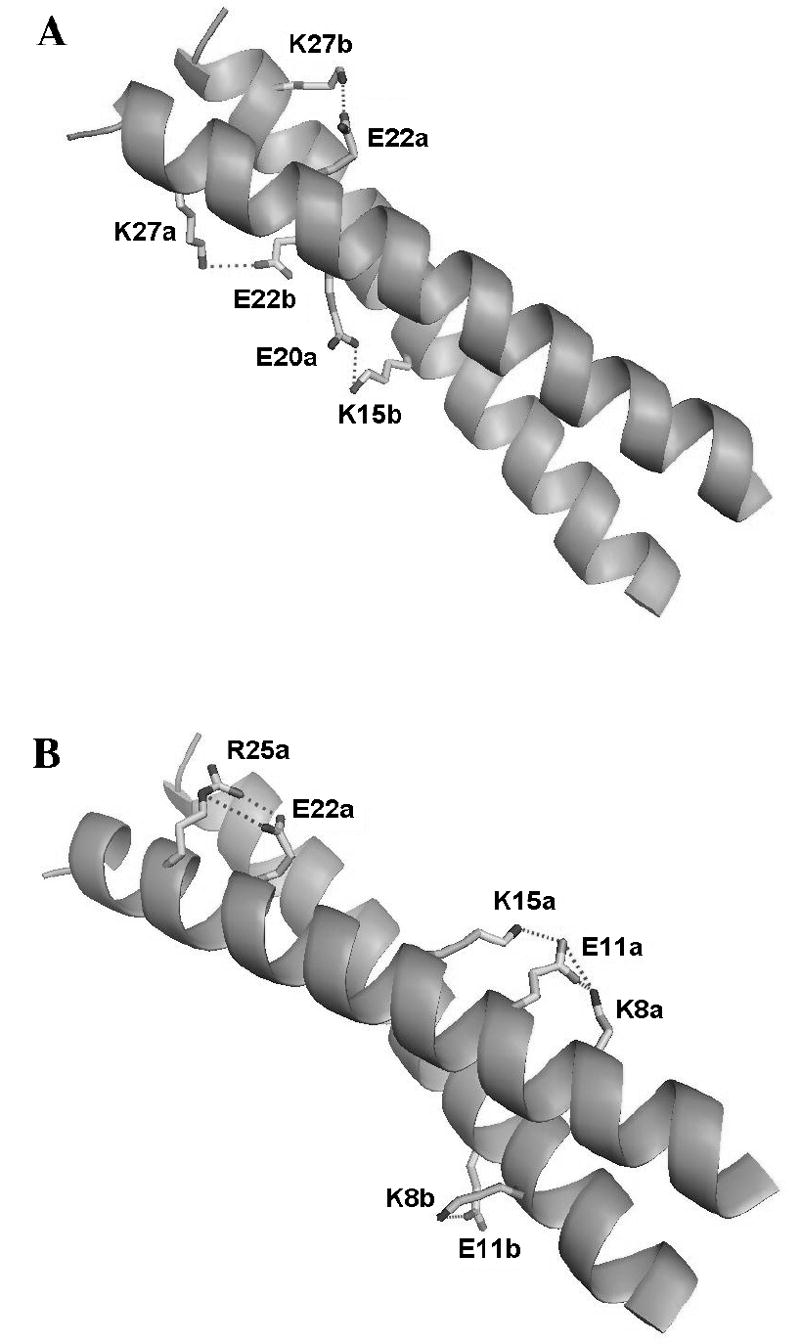
Diagram of the GCN4p leucine zipper structure (PDB code 2zta.pdb) showing (A) inter-chain and (B) intra-chain ion pairs. The designations “a” and “b” refer to the two monomers of the coiled coil homo-dimer. Note that some of the ion pairs are only observed for one of the two monomers of the homodimer in the crystal structure (Table 2).
NMR provides direct information on the contributions of individual ionizable groups to protein stability. In a pH titration experiment, resonances from nuclei in the vicinity of an ionizable residue experience gradual chemical shift changes dependent on the ratio of the group’s conjugate acid and base forms. The microscopic pKa is obtained from the pH at which the chemical shift changes between acidic and basic extremes are half-maximal 27.
To date, NMR investigations have focused almost exclusively on residues that titrate near neutral or acidic pH. The ionization of basic residues is more difficult to study by NMR owing to the loss of amide proton resonances through fast hydrogen exchange at high pH. The availability of 13C/15N double-labeled samples in conjunction with an alternative 1H-13C HSQC approach, made it possible to determine the pKa values for all 16 ionizable groups in the 33-residue leucine zipper fragment that titrate in the pH range 1 to 13. Besides determining the pKa values for the N state, we determined pKa values for the D state in 6 M urea, and for two peptide fragments (p2-16 and p16-31) intended to mimic the D state of the protein in the absence of denaturants. This is one of only a few investigations to obtain pKa data for the D state 28-30 and possibly the first to characterize both acidic and basic residues. Thermodynamic linkage analysis 17,31 of the differences in pKa values between the N and D states for individual ionizable groups, affords a comprehensive assessment of electrostatic contributions to the stability of GCN4p. The results are analyzed in the context of available X-ray structures of the leucine zipper, mutagenesis data, urea unfolding experiments, and structure-based continuum calculations of pKa values.
Results
pKa values in the native state
To obtain data for as many ionizable groups as possible, we recorded 1D and 2D NMR experiments at over 30 intervals between pH 1 and 13. The pH titrations of aliphatic residues were characterized using sensitivity-enhanced 2D 1H-13C HSQC experiments, which were run in constant time mode and were selective for -CH2- groups 32. Representative data at three pH values are shown in Fig. 2. Titrations were followed using the 1H and 13C chemical shifts of the methylene group closest to the ionizable moiety. We opted for this approach rather than the alternative side-chain H(CA)CO experiment 22,32,33 because the latter suffers from poor sensitivity, and is limited to acidic residues. The pKa values of aromatic residues were obtained from 1D 13C-edited NMR spectra with the carbon transmitter set in the aromatic region. Additional 2D 1H-15N HSQC experiments on the same samples supplemented the carbon NMR data. To follow crossovers of side-chain resonance positions with pH, we recorded 3D HCCH data sets at pH 2.5, 4.5, 5.8, 7.4 and 12.5, and 3D 13C-edited NOESY-HSQC data at pH 2.5, 6.9 and 10.5. These pH values correspond roughly to the mid- and end-points of the pH titrations for acidic and basic residues. All pH data were obtained in 90% H2O/10% D2O to avoid any possible corrections for the deuterium isotope, at a temperature of at 25 °C, and at a physiological salt concentration of 150 mM NaCl.
Figure 2.
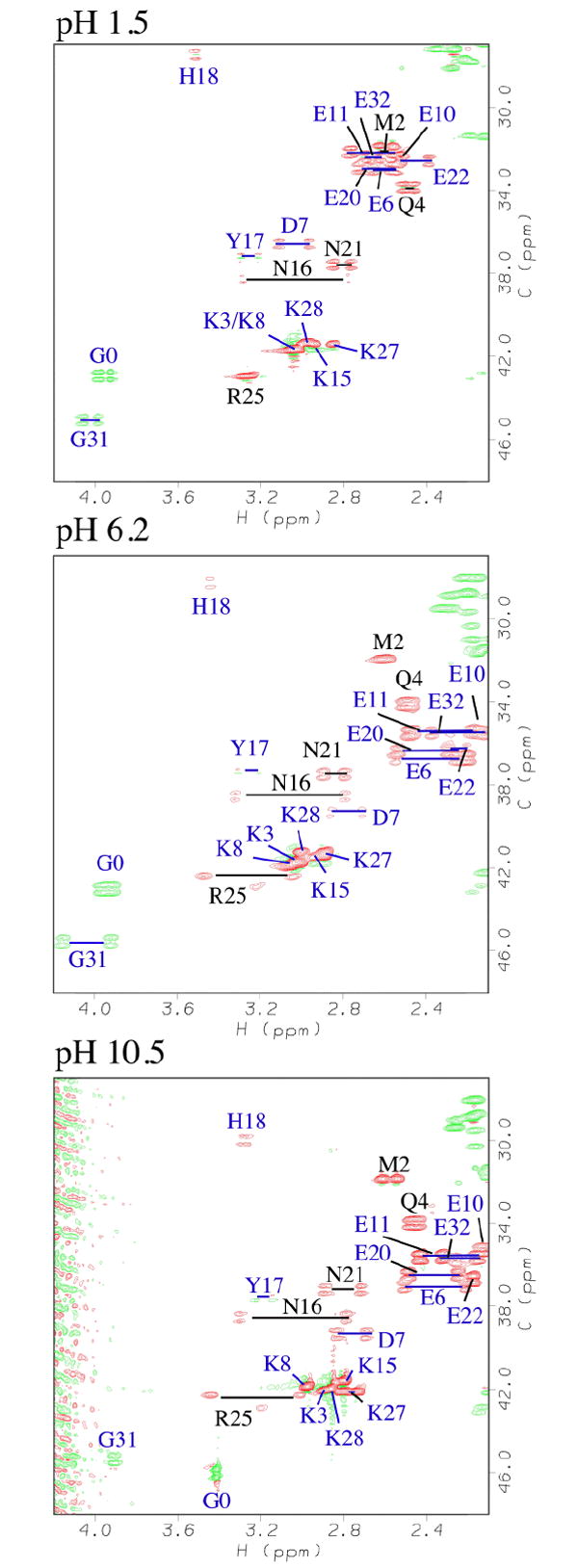
Representative constant-time 2D 1H-13C HSQC spectra 32 used to characterize the titration of native GCN4p. Residues are labeled according to position in the sequence. Horizontal bars connect non-degenerate methylene protons. In constant time mode with T = 1/JCC, carbons coupled to an odd number of aliphatic carbons, give opposite phases from those coupled to an even number 32. Positive and negative phases are represented in the figure by red and green contours.
Figure 3 shows representative pH titration data fit to a modified Henderson-Hasselbalch equation 27. The pKa values obtained from the fits are given in Table 1. The N-state pKa values are tightly clustered within a range of ~0.6 pH units for residues of the same type. The average difference from random coil values 34 is 0.3 pH units, and the largest is 0.86 pH units for Lys8.
Figure 3.
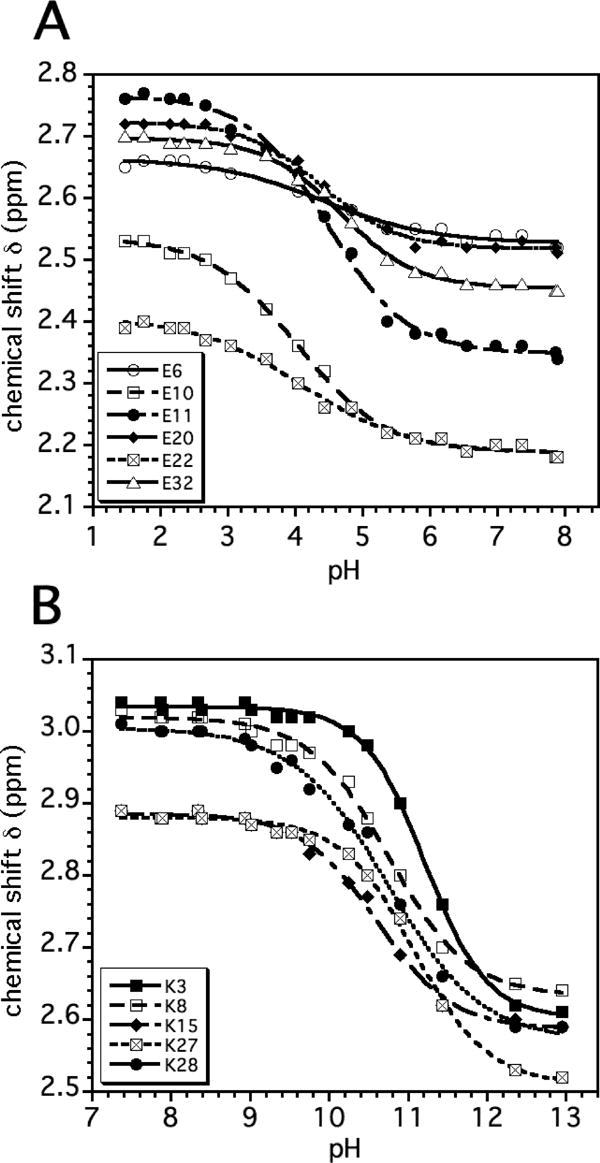
Representative titration data showing (A) the six glutamates and (B) the five lysines in the N-state. The curves are least-squares fits of the data to a modified Henderson-Hasselbalch equation 27,64. The pKa values from the fits are given in Table 1 and titration parameters in the Supplementary Tables 1-3.
Table 1.
Summary of pKa values and contributions of charges to GCN4p stability. a
| Group | pKa in N state | pKa in D state (6 M urea) b | pKa in D state (peptides) c | pKa for random coild | ??Gurea – N (kcal/mol dimer) e | ??Gpept – N (kcal/mol-dimer) f |
|---|---|---|---|---|---|---|
| N-termi | 7.88 ± 0.05 | 7.74 ± 0.05 | N.D. | 7.5 | -0.38 ± 0.19 | N.D. |
| Lys 3 | 10.78 ± 0.05 | 10.49 ± 0.15g | 10.56 ± 0.08g | 10.4 | -0.79 ± 0.19 | -0.60 ± 0.26 |
| Glu 6 | 4.60 ± 0.15 | 4.11 ± 0.20 | 4.08 ± 0.06 | 4.25 | 1.33 ± 0.68 | 1.42 ± 0.44 |
| Asp 7 | 3.48 ± 0.05 | 3.65 ± 0.05 | 3.55 ± 0.08 | 3.67 | -0.46 ± 0.19 | -0.19 ± 0.26 |
| Lys 8 | 11.26 ± 0.05 | 10.49 ± 0.15g | 10.56 ± 0.08g | 10.4 | -2.10 ± 0.19 | -1.91 ± 0.26 |
| Glu 10 | 3.94 ± 0.10 | 4.20 ± 0.16 | 4.17 ± 0.13 | 4.25 | -0.71 ± 0.51 | -0.63 ± 0.45 |
| Glu 11 | 4.05 ± 0.05 | 4.41 ± 0.05 | 4.40 ± 0.05 | 4.25 | -0.98 ± 0.19 | -0.95 ± 0.19 |
| Lys 15 | 10.62 ± 0.06 | 10.49 ± 0.15g | 10.56 ± 0.08g | 10.4 | -0.35 ± 0.21 | -0.16 ± 0.27 |
| Tyr 17 | 9.82 ± 0.08 | 10.14 ± 0.05 | 9.86 ± 0.05 | 9.84 | -0.87 ± 0.26 | -0.11 ± 0.26 |
| His 18 | 6.24 ± 0.06 | 6.38 ± 0.05 | 6.64 ± 0.06 | 6.54 | 0.38 ± 0.21 | 1.09 ± 0.23 |
| Glu 20 | 4.38 ± 0.05 | 4.03 ± 0.05 | 4.12 ± 0.05h | 4.25 | 0.95 ± 0.19 | 0.71 ± 0.19 |
| Glu 22 | 4.20 ± 0.08 | 4.01 ± 0.05 | 4.20 ± 0.05h | 4.25 | 0.51 ± 0.26 | 0.00 ± 0.09 |
| Arg 25 | > 13.0 | > 13.0 | > 13.0 | 12.0 | N.D. | N.D. |
| Lys 27 | 11.10 ± 0.05 | 10.49 ± 0.15g | 10.56 ± 0.08g | 10.4 | -1.66 ± 0.19 | -1.47 ± 0.26 |
| Lys 28 | 10.64 ± 0.12 | 10.49 ± 0.15g | 10.56 ± 0.08g | 10.4 | -0.41 ± 0.35 | -0.22 ± 0.39 |
| Glu 32i | 4.62 ± 0.11 | 4.50 ± 0.09 | N.D. | 4.25 | 0.33 ± 0.39 | N.D. |
| C-termi | 4.03 ± 0.06 | 3.62 ± 0.07 | N.D. | 3.67 | 1.12 ± 0.25 | N.D. |
All data were obtained at a temperature of 25 °C. The errors reported for the pKa values are the largest of the following numbers: the standard error of the fit in cases where only one resonance was reliable for following the titrating group, the standard error of the mean in cases where multiple resonances could be used follow a titrating group (see Supplementary tables 1-3), and 0.05 pH units which is likely to be the accuracy limit of the electrodes used to measure pH.
All values in this column include a −0.52 pH unit correction for the effect of urea on the pH electrode.
D-state values obtained from the overlapping peptides p2-16 and p16-31 in the absence of denaturants.
Random coil model compound values 34.
??G values using the urea-denatured protein as a model for the D-state.
??G values using the overlapping peptide fragments p2-16 + p16-31 as models for the D-state.
The lysine Cε-Hε crosspeaks titrated as an unresolved group in the D state. For the urea-denatured state we obtained a pKa of 10.49; for the C-terminal p16-p31 peptide fragment 10.48; and for the N-terminal p2-16 peptide fragment 10.65. The difference between the pKa values of lysines from the N- and C-terminal peptides probably reflects experimental error. For the peptide fragment model of the denatured state, we therefore used the averaged pKa value of 10.56.
Data from a p15-27 peptide fragment at 25 °C and 150 mM NaCl 17.
The N-terminus of the molecule is Gly0. This residue is an artifact of the engineered thrombin cleavage site used to purify the protein 26. The C-terminus of the molecule is Glu32. The pKa value for Glu32 is for the side-chain. The pKa of the Glu32 α-carboxylic acid group was measured from Gly31 Hα in the N state and from E32 HN in the D state, and is indicated with the label ‘C-term’ in the table.
pKa values in the denatured state
Ionization constants of residues in unfolded proteins can be affected by nearby residues in the sequence, by the intrinsic hydrophobicity of the polypeptide, and by residual structure still present under denaturing conditions. For these reasons, pKa values obtained directly from the D state are more accurate than those inferred from random coil model compounds 12,28,29,35. To model the D state of GCN4p we studied the protein in 6 M urea, since this is often the endpoint of protein denaturation studies 13. In addition we looked at two overlapping unfolded peptide fragments (p2-16 and p16-31) that encompass the sequence of GCN4p. The rationale for studying these fragments is that they allowed us to obtain D-state pKa values in the absence of denaturants, thereby circumventing effects specific to the denaturant, effects of the denaturant on pH, and effects of the denaturant on the pH electrode (see below). Full-length GCN4p retains about 5-10% residual α-helix structure in 6 M urea (A.T.A. & W.M., in preparation). The p16-31 fragment has a fractional population of ~50% helix at 5 °C in water but less than 25% α-helix at 25 °C 36,37. These levels of residual structure should not introduce large errors in the population-averaged pKa values for the D state. Moreover, the structure that persists in these systems is representative of the initial states for folding of the coiled coil, in urea or water.
The pKa values for the D state were measured with the same methods as for the N state. The chemical shift dispersion of the six Glu residues in 2D 1H-13C HSQC of the D state was very poor. To overcome this, we followed the titrations of the Glu residues from their backbone amide protons in 1H-15N HSQC spectra. Proteins have inherently good 15N dispersion even when unfolded, and the titration of residues more than one or two positions in the sequence has negligible effects in denatured proteins 38. We could not use 1H-15N HSQC spectra to characterize the lysine residues because of fast amide proton exchange at high pH. We determined a ‘group pKa’ of 10.5 from a single unresolved Hε-Cε methylene crosspeak in 2D 1H-13C HSQC spectra, and that the pKa values of individual lysines in the D state were with ~0.15 pH units of this average for the urea-denatured state. For the peptide model we obtained an average lysine-group pKa of 10.6
The pKa values for the D states of GCN4p are given in Table 1. For the urea-denatured state it was necessary to subtract 0.52 pH units from the measured pKa values, to correct for the effects of urea 31,39. This correction does not distinguish between the effect of urea on the pKa, and the effects of urea on the pH electrode40. The correction was based on experiments with the model compound N(α)-acetyl-L-histidine31 (not shown) and was assumed to be applicable over the entire pH range studied. Corrections for the effects of urea on pH such as the ones used here, were found to vary little with pH, at most 0.2 pH units for groups that titrate between pH 4.1 and 10.339. To further rule out possible problems associated with the correction for the effects of urea on the apparent pH, we collected a second set of D-state pKa values using the unfolded p2-16 and p16-31 peptide fragments of GCN4p in water. The agreement between the pKa values obtained in 6 M urea, and for the peptide fragments in water (Table 1) supports the validity of the correction used for urea.
The pKa values obtained for the D state of GCN4p in urea, for the peptide fragments in water, and literature values for putative random coil peptides 34 are very similar, with an R.M.S difference of ~0.15 pH units between the three data sets. We estimate that the uncertainty of our pKa measurements is at least 0.05 pH units (Table 1), since this on the order of the accuracy of the electrodes used to measure the solution pH. Previous studies found larger differences between the pKa values of D-states and random-coil model compounds 28-30 but the differences were suppressed at high ionic strength 29,30. The better agreement in the present study is probably due to the use of a relatively high salt concentration (150 mM NaCl) and the use of updated random coil values 34. In spite of the overall agreement, there are some substantial individual differences. Tyr17 for example, has a pKa that is 0.3 pH units higher in 6 M urea than in water (Table 1) possibly as a result of solvation differences for the phenolate ion under the two sets of conditions. These subtle effects highlight the need to measure pKa values directly from the D state.
Contributions of ionization equilibria to coiled coil stability
The shifts in pKa values accompanying protein folding are related to the contribution of a change in charge to protein stability by 17,31:
| (1) |
Equation 1 is used as is in the case of acidic groups 17 but needs to be multiplied by −1 for basic groups 1,31. For a dimer, Equation 1 needs to be multiplied by a factor of 2 to account for the contributions from each of the magnetically equivalent subunits. Values for ΔΔGtitr calculated from differences in pKa shifts of individual groups between the N and D states are given in Table 1, and contributions greater than 0.9 kcal/mol are highlighted in bold text.
We were able to determine pKa values for all ionizable residues except Arg25. The Cδ methylene resonances of Arg25 do not appear to shift above pH 8. Instead, a new resonance appears at ~pH 12 at the expense of the resonances seen at lower pH, suggesting that the charged and uncharged forms of Arg25 are in slow exchange on the NMR timescale. All other ionizable groups in the protein show pH titrations that are in n the fast exchange regime on the NMR timescale. Both the folded and urea-unfolded states of GCN4p show the same behavior for Arg25. Unfortunately, the titration for Arg25 is not complete even at pH 13.3, the highest pH studied. We could therefore not determine the pKa of Arg25 but conclude that it is higher than pH 13 in both N and D states. Mutagenesis experiments strongly suggest that the charged form of Arg25 is involved in a favorable interaction with Glu22 that stabilizes the N-state by over 2 kcal/mol 13,26. Electrostatic interactions between Arg25 and Glu22 also play a critical role in the kinetics of coiled coil folding, by stabilizing a nascent α-helical nucleation site within the trigger sequence of GCN4p 36,37.
Structural characterization of interactions between charged residues
Negative contributions in Table 1 imply energetically favorable electrostatic interactions. A large number of hydrogen-bonded ion pairs (salt bridges) are found in the X-ray structure of GCN4p (Fig. 1), yet only some of these correlate with significant favorable contributions to stability (Table 1). We used 13C-edited 3D NOESY-HSQC data (Supplementary Materials Figure 1) to test for distance contacts that could support or refute the presence of the ion pairs observed in the X-ray structure. NOEs were observed between Glu22(Hγ)-Arg25(Hδ), Glu22(Hγ)-Lys27(Hε), and Lys8(Hγ)-Glu11(Hγ) at pH 6.9; consistent with the presence of the three salt-bridges in the X-ray structure, and consistent with the favorable energetic contributions from the charged forms of residues Lys8, Glu11, and Lys27 (Table 1). The NOEs corresponding to the ion pairing interactions observed at neutral pH disappear at pH 2.5. This indicates that the NOEs are due to charge-charge interactions rather than structural features independent of pH. We failed to detect NOEs to support the Glu11-Lys15 and Lys15-Glu20 salt-bridges, which are only seen in the X-ray structure for one monomer of the dimer (Fig. 1, Table 2). Moreover, the pKa values of Lys15 and Glu20 do not suggest stabilizing interactions (Table 1) but rather that the charged form of Glu20 is destabilizing17.
Table 2.
Distances of potential ion pairs in the X-ray structure of GCN4p.
| Interaction | d(chain a)
(Å) |
d(chain b)
(Å) |
<dwild type> a (Å) |
<dfamily> b (Å) |
|---|---|---|---|---|
| Lys8 – Glu11 | 3.3 | 5.0 | 4.1 | 4.3 |
| Glu11 – Lys15 | 4.6 | 10.2 | 7.4 | 5.8 |
| Lys15 – Glu20 | 3.3 | 6.2 | 4.8 | 6.7 |
| Glu22 - Arg25 | 2.8 | 4.6 | 3.7 | 4.5 |
| Glu22 - Lys27 | 3.6 | 3.3 | 3.4 | 3.8 |
| Asp7 – Glu10c | 3.5 | 5.6 | 4.6 | 4.8 |
| Arg25 – Lys27c | 3.7 | 7.7 | 5.7 | 6.7 |
Distance average of the values for chains a and b of the homodimer.
Average over a family of GCN4p X-ray structures (PDB codes: 2ZTA, 1ZII, 2AHP, 1GK6, 1KQL, 1ZIK, 1ZIL) that includes distances for both chains a and b of each mutant structure.
Potential repulsive interaction between like charges.
Information on the proximity of ionizable residues can also be obtained from secondary pH titrations that manifest themselves in the high or low pH chemical shift plateaus of the oppositely charged ion pair partner 27,35. The titration of Glu22 is reflected in the resonances of both Arg25 and Lys27, consistent with the Glu22-Arg25 and Glu22-Lys27 ion pairs observed in the X-ray structure of GCN4p (Fig. 1), and with the NOE data described above. We also see a weak reflected titration for Glu11 consistent with the Glu11-Lys8 ion pair. Additional secondary titrations are seen for residues Glu6, Asp7, Glu20 that can be assigned to the ionization of nearest neighbors in the sequence. The secondary titrations span chemical shift ranges of ~0.05 ppm for 1H, compared to ~0.3 ppm for the primary titrations making it difficult to obtain accurate pKa values.
pH dependence of the free energy for unfolding
The role of electrostatic interactions was further examined with far-UV CD experiments monitoring the urea-induced unfolding of GCN4p in the range between pH 2 and 12 (Figures 4, 5 and Supplementary Table 4). The denaturation curves were used to extract ΔG0 and m-values by the linear extrapolation method (Fig. 5, Supplementary Table 4), which will simultaneously correct for the effects of the denaturant on ΔG, and for the effects of the denaturant on pH41. The stability of the coiled coil expressed in terms of ΔG0 values is relatively constant between pH 4 and 10, and only decreases at the extremes of pH (Fig. 5). There is a small dip in stability profile near pH 5 (Fig. 5), which coincides with the appearance of a minor form of the protein in 1H-15N HSQC spectra (not shown). This minor form, which has been described in detail in a publication42 that appeared while this paper was under review, is populated at levels below 10% under the conditions of our NMR studies. The low concentration of the minor form, and the fact that the dip in the pH profile is reproduced in stability profiles calculated from the GCN4p X-ray structure that did not take the minor form into account, make it unlikely that it is linked to the dip in the stability profile.
Figure 4.
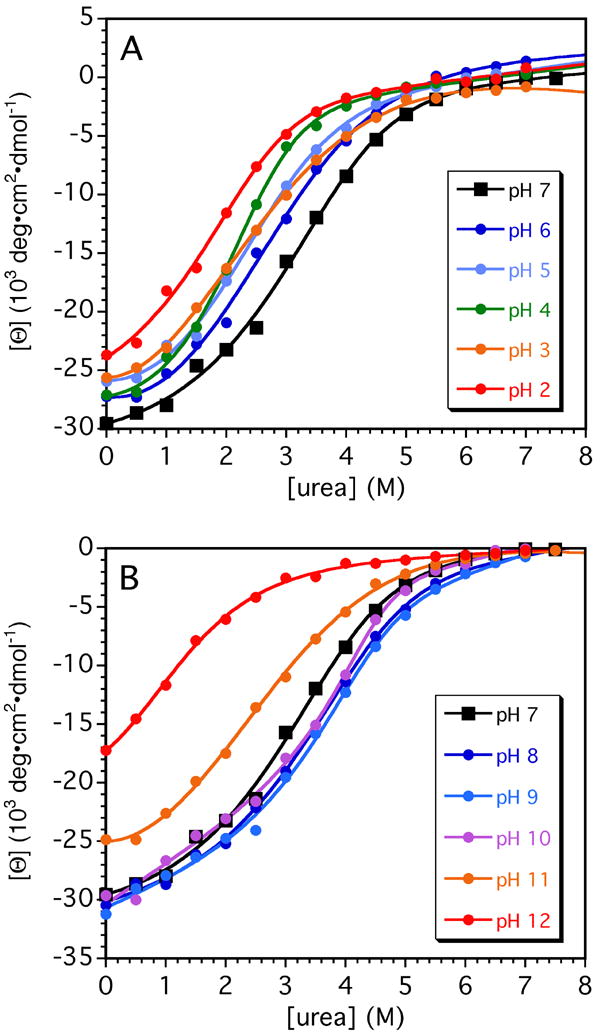
Urea denaturation curves of GCN4p as a function of pH. Denaturation transitions were measured by circular dichroism at Θ222, and were fitted to Equation 3 to determine ΔG0 and m-values at each pH. The parameters describing the stability of the coiled coil are summarized in Supplementary Table 4. The inset gives the color-coding used to represent the different pH values.
Figure 5.
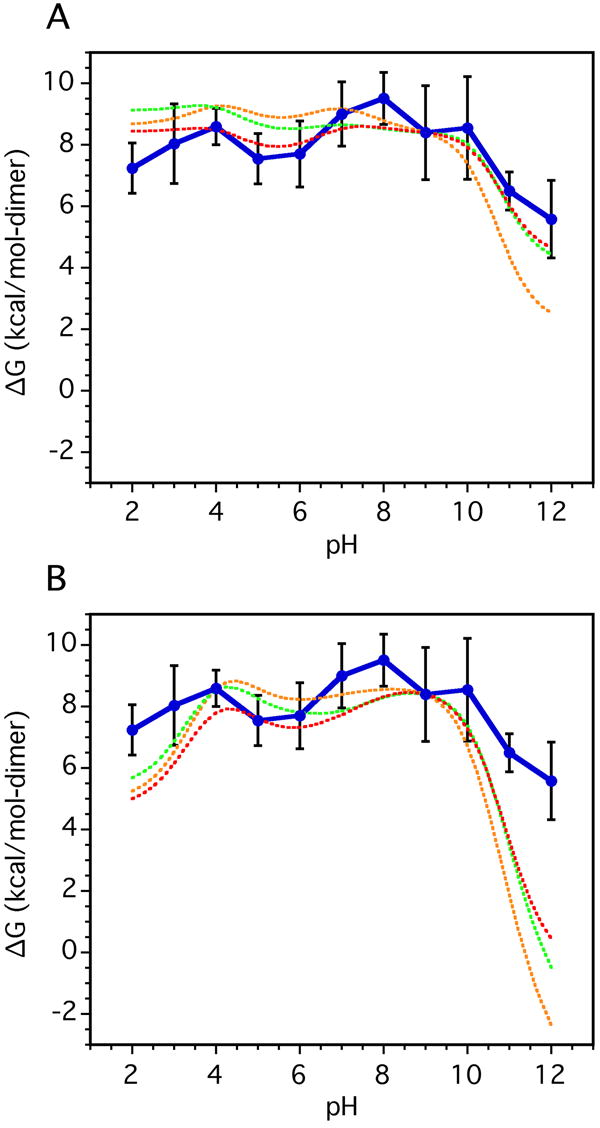
Charge contributions to the pH dependence of GCN4p stability. In both panels the solid lines with error bars represent the experimental urea-unfolding data from Fig. 4; the different dashed curves represent calculations assuming different pKa values for the U state. The energies calculated are not absolute free energies therefore for comparison the simulated curves were arbitrarily superimposed to the value of the experimental curve at pH 9. (A) Stability estimates obtained by numerical integration of the difference in H+ binding curves calculated from the pKa values of the N and D states. Different models were tested for the D-state: green - pKa values measured for GCN4 in urea, red - pKa values determined in water from the overlapping p2-16 and p16-31 peptide fragments that encompass the GCN4p sequence (see Table 1), orange - pKa values from potentiometric titrations of short model peptides 34. (B) Same as above, except using theoretical N state pKa’s calculated with the FDBP approach from the GCN4p X-ray structure in lieu of those measured by NMR.
The coiled coil is destabilized by ~2 and 4 kcal/mol at the extremes of acid and basic pH, compared to the stability maximum of 9.5 kcal/mol at pH 8 (Fig. 5). The relative insensitivity of the ΔGunf profile to changes in pH is consistent with the small differences in pKa values between the N and D states. Hydrophobic and hydrogen bonding interactions are the principal forces stabilizing the GCN4p coiled coil. Nevertheless, the decrease of ~4 kcal/mol between pH 8 and pH 12 when all other variables are held constant, indicates that electrostatics also play a significant role in stabilizing the N state of GCN4p. The asymmetry of the pH profile is consistent with larger pKa shifts between D and N states for basic compared to acidic groups (see Table 1).
Theoretical and structure-based analysis of the pH dependence of stability
The pH dependence of stability data obtained from the urea unfolding experiments (Table 2) is compared in Figure 5a to stability profiles calculated 43 by numerical integration of the H+ binding curves derived from the pKa data (Table 1). The experimental data are shown with a solid line; the different dashed curves represent calculations assuming different pKa values for the U state. All the modeled curves in this figure were superimposed arbitrarily to the value of the experimental curve at pH 9. The differences between the predicted stability curves illustrate the high sensitivity of the numerical integrations to small differences in D state pKa values 44-46, which approach the experimental uncertainty of ~0.1 pH units (Table 1). This sensitivity further underscores the need for an improved understanding of the electrostatic properties of D states.
The agreement between the stability profiles measured from equilibrium denaturation and estimated from pKa values is excellent between pH 3 and pH 10 (Fig. 5a). The shapes of the profiles agree well with the equilibrium unfolding data, including the asymmetry where alkaline pH is more destabilizing than acidic pH, and the local dip between pH 5 and 6. Below pH 3 the agreement is slightly better when the peptide data in water (red) are used in lieu of the data collected for the protein in 6M urea data (green). Above pH 10 all of the models underestimate the stability measured from denaturation experiments by ~1 to 3 kcal/mol, with the random coil values (orange) showing the largest differences. The underestimation is likely to reflect the lack of a measured pKa value for Arg25, which based on mutagenesis data stabilizes the protein by 2 kcal mol 13,26.
Figure 5b shows stability versus pH curves obtained by replacing the N state pKa values measured by NMR with values predicted from the X-ray structure of GCN4p (PDB accession code 2ZTA) using continuum electrostatic calculations. The calculations are based on the numerical solution of the linearized Poisson-Boltzmann equation, as described in the Methods section. As seen in Fig. 5b the calculated stability curves match the experimental profile quite well through the range between pH 4 and 10. At the extremes of pH outside this range, the structure-based simulations severely overestimate the contributions of electrostatic interactions to the stability of the coiled coil. Overestimation of electrostatic effects in structure-based calculations with continuum methods is a common problem 4,46. Presumably this arises from local flexibility and small conformational reorganization experienced by the protein in solution, which is not reproduced in the calculations with static structures. The calculations attempt to reproduce these effects implicitly by using arbitrarily high protein dielectric constants, but evidently, this is not sufficient to capture the solution behavior of proteins under extreme conditions 46,47.
Discussion
The contributions of electrostatic interactions to protein stability have generated considerable interest due to their importance in protein folding, modeling, and design. There has also been controversy for coiled coils in particular, surrounding the apparent disagreement of results obtained with different experimental methods 1,11-13,17. The availability of a complete pKa data set for the GCN4 leucine zipper puts us in a position to try to reconcile some of the apparent discrepancies.
Are ion pair propensities different in crystals and in solution?
Ion pairs appear to be more common in X-ray structures of proteins, than in solution 13,17. Several factors could contribute: 1) Crystal structures are typically done at lower temperatures than solution NMR studies (as low as 100 °K for cryo-crystallography). The pKa values for GCN4 were measured at 25 °C, whereas the x-ray structure was determined at 5 °C (T. Alber, personal communication). Although ionization enthalpies are typically small, these can result in pKa shifts that are significant compared to the small differences in pKa values between N and D states 48. Low temperatures could also induce structuring of surface residues directly by reducing motion, or indirectly by affecting the properties of the water in the crystal 49. (2) Electrostatic interactions are modulated by the dielectric effect 2,50,51. It is much less clear how the dielectric effect for a protein in solution compares to that in the crystalline state. Differences in counter-ion concentrations in solution and in crystals could play additional roles 52, as would osmotic effects associated with the precipitants and cryoprotectants used to grow crystals. We are aware of only one study using the protein ribonuclease A that attempted to correlate pKa values from solution NMR 53 with those obtained in the crystalline state using X-ray diffraction 54. The pKa of ribonuclease A residue His119, which is not involved in a salt-bridge was identical in the crystal and solution 53,54. His12 and His48 which participate in ion pairs in the X-ray structure both had their pKa values raised from pH 6.0 in solution to pH 7.0 in the crystal 53,54, suggesting that the ion pairs are more difficult to break in the crystal. (3) The structures of surface residues in crystals can be subject to distortions from lattice packing. These account for differences in structure between the two GCN4p monomers (Fig. 1), which NMR shows are magnetically equivalent in solution, and probably also accounts for some of the differences between closely related X-ray structures of single site GCN4p mutants. Table 2 summarizes the distances between the closest charged side-chain N and O atoms for the potential ion pairs in GCN4p (Fig. 1). The cut-off distance typically used in the literature to identify an ion pair is 4 Å between the side-chain N and O atoms of oppositely charged residues 55. If the averages of the distances for symmetry-related monomers are examined, the interactions with the shortest average ion-pair distances correspond with the residues that have stabilizing ΔΔG contributions (Table 1). The same trend is seen if we average over all the distances in a family of seven structures closely related to GCN4p. Note that only the Glu22-Lys27 interaction strictly satisfies the 4 Å threshold for all the structures (Table 2). These observations indicate that distances averaged over a family of X-ray structures are more representative of the ion-pairs found in solution.
Non-additive charging effects for partners in ion pairs
Discrepancies between X-ray and NMR results on the contributions of charges to protein stability were first noted for Glu20 and Glu22 of GCN4p 17. The two glutamates form Glu22-Arg25 and Glu22-Lys27 salt-bridges in the X-ray structure 15 but showed only small, and slightly unfavorable contributions to stability by NMR 17, which we confirm in the present work (Table 1). Our more complete pKa data set shows that if we consider the basic residues one of the ion pair partners, Lys27, makes a large favorable contribution of -1.7 kcal/mol to the N state (Table 1). The titration of the second ion pair partner, Arg25, could not be determined but mutagenesis data 13,26 and the larger pKa for Arg25 than arginine model compounds suggests the charged form of Arg25 also makes favorable contribution to stability. For the remaining Lys8-Glu11 salt bridge supported by NOEs, both residues give favorable contributions, with that for Glu11 twice as large as for Lys8.
These results emphasize that the ΔΔGurea – water values for partners across ion pairs need not be correlated. Discrepancies between pKa values of interacting charged residues have been reported in a number of systems 31,53,56. In one example of a reversed ion pair, it was shown that the contribution to the stability of the protein depended on the orientation of the ion pair 57. The ionization of a residue in an ion pair can contain energetic terms in addition to the ion pair contribution 1. These include contributions associated with the desolvation of charged compared to uncharged groups; and residual interactions, for example if hydrogen bonding is retained between the charged and uncharged versions of the interacting residues 1. The context provided by the medium and long-range interactions are as important as the ion pair itself. The pKa shift determines the free energy change for ionizing a residue in the N compared to the D state, but does not include contributions for the unsatisfied ion pair partner 13, which need to be measured separately.
Comparison of NMR and mutagenesis results
Substitution of charged residues with neutral counterparts often destabilize coiled coils, whereas NMR studies show smaller effects 11,12,17. The Matthews group published 13 comprehensive data on the stability effects of mutations involving charged residues in GCN4p, which we can now compare with our complete pKa data set. The published ΔΔGwt-mut values for single-site mutants that replace charged residues in GCN4 13 are weakly correlated with the corresponding ΔGurea – H2O values obtained from NMR in the present study. The only glaring discrepancy occurs for Glu22 where mutagenesis predicts a large destabilizing effect when the charge is removed, whereas NMR predicts a small stabilizing effect. Glu22 is unusual (Fig. 1, Table 2) in that it simultaneously participates in two hydrogen-bonded ion pairs (salt bridges): Glu22-Arg25 and Glu22-Lys27’. An analysis in terms of two isolated charges may be thus be inappropriate 13,15,26. Excluding Glu22, the NMR and mutagenesis data are weakly correlated with an R-value = 0.78, ρ = 0.07 for n = 6 comparisons. The number of comparisons is too small for good statistics, and would benefit from data on additional proteins.
Comparison between experimental and theoretical pH-stability profiles
There is reasonable agreement between the pH dependence of stability calculated by integration of H+ binding data obtained from NMR-derived pKa values, and the stability measured from urea denaturation experiments of GCN4p (Fig. 5a). In the basic pH region the numerical integration curves predict a larger drop in stability than observed, but the discrepancy may be due to the fact that there are no data for Arg25 which is known from mutagenesis to stabilize the N state by about 2 kcal/mol•dimer. The equilibrium unfolding data (Fig. 4, Table 2) indicate that starting from the stability maximum of 9.5 kcal/mol at pH 8, the leucine zipper loses about 2 kcal/mol•dimer and 4 kcal/mol•dimer when the protein is brought to the extremes of acidic and basic pH, respectively. The stabilities derived from pKa measurements predict comparable losses of about 1 kcal/mol•dimer and 5 kcal/mol•dimer from pH 8 to the extremes of acidic and basic pH (Fig. 5a).
Based on the equilibrium unfolding data for GCN4p at a 30 μM protein concentration, electrostatic effects contribute as much as 20-40% of the stability decrease of GCN4p going from pH 9 to 12. In addition to hydrophobic effects, van der Waals packing, and hydrogen-bonding, charge-charge interactions thus also clearly stabilize the coiled coil. A limitation of pKa investigations by NMR spectroscopy is that the protein must remain folded over the range of pH values studied. This has the potential to bias results against a system in which ion pairs make large contributions to stability. The exceptional stability of GCN4p, which enabled the protein to be studied between pH 1 and 13 is atypical of the majority of proteins that unfold at much milder pH values. For these proteins, electrostatic contributions may be larger but inaccessible because the N state is too destabilized to study over a large pH range.
In simulations where the experimental N state pKa values are replaced with ones predicted from the X-ray structure, the shape of the pH profile was reproduced but the simulations over-predicted the magnitude of electrostatic contributions to stability at the extremes of pH (Fig. 5b). The simulations suggest that the coiled coil should be predominantly unfolded above pH 11. This was not observed experimentally. The overestimate probably reflects the higher propensity for ion pairs to form in crystals compared to solution, and the inability of continuum calculations to treat explicitly the structural reorganizations that occur at the extremes of the pH range.
Prospects for improved modeling
In spite of considerable progress towards the calculation of pKa values from structure, some fundamental aspects of pKa modeling remain unresolved. Structural modeling of protein electrostatics is based in most cases on X-ray structures. There is currently very little information on how surface charges behave in crystals compared to solution. Differences in the effective and apparent dielectric effects, in counter-ion concentrations, and the reduction of side-chain motion in the crystalline state could all play roles in favoring ion pairs in crystals, compared to solution. Comparative studies of the type done with ribonuclease A 54, would help determine the extent to which charges behave the same way in crystals and in solution. Crystal lattice packing can distort the structures of protein surfaces as exemplified by the poor conservation of the GCN4p ion pairs when different X-ray structures are compared (Table 2). When available, a family of X-ray or NMR structures could improve modeling by distinguishing invariant interactions from structural noise. Another approach makes use of MD simulations to model dynamics starting from X-ray structures 58-60. We performed pKa calculations using the continuum methods with GCN4 structures that were relaxed for 10ns in MD simulations, but this did not improve the overall quality of the agreement between the calculated and measured pKa values or the calculations of the pH dependence of stability.
The most important open question in improving the accuracy of pKa calculations is the nature of structural reorganizations accompanying changes in pH. Backbone as well as side-chain NMR resonances of non-titrating residues are well conserved in spectra of GCN4p over the entire pH range studied (Fig. 2), indicating that the overall structure is preserved. However, subtle changes in structure or dynamics do occur, and appear to be localized to residues that participate in ion pairs, as demonstrated by changes in NOE contacts for these residues with changing pH (Supplementary Figure 1). The characterization of these rearrangements is likely to be critically important for accurate modeling of pKa values, and is a challenge for future experimental and computational studies.
Materials and Methods
Sample preparation
A synthetic gene encoding residues Met250 through Glu280 of GCN4 (NCBI accession number NP_010907) was prepared for optimal codon usage in Escherichia coli 37, and ligated into the BamHI/EcoRI site of the pHisTrx2 plasmid 61, a derivative of pET-32a (Novagen). The plasmid was used to transform E. coli JM109(DE3) cells (Promega), which were grown overnight in 100 mL of LB medium, collected by centrifugation at 500-1000g (25°C), and resuspended in 1 L of ‘new minimal medium’ 62 supplemented with 1 g/L 15NH4Cl and 3 g/L 13C6-glucose (Spectra Stable Isotopes). When the cells growing at 37 °C reached an OD600 of 0.6, expression of the His6-thioredoxin-GCN4p fusion protein was induced with 1 mM IPTG. The induced culture was allowed to grow for an additional 4 h at 37 °C before harvesting by centrifugation at 5000g. The resulting pellet was sonicated (6 × 30s), and applied to a Ni2+-Sepharose (Novagen) column under denaturing conditions. Thrombin cleavage of the recombinant GCN4p fragment from the 6xHis-tagged thioredoxin carrier protein was performed as described in the manufacturer’s instructions (Amersham). Before use, the purified GCN4p-wt protein was dialyzed extensively against 10 mM NaCl, 10 mM sodium phosphate, pH 6.2, using 3.5 kDa cutoff Biotech dialysis tubing (SpectraPor). Samples were lyophilized and stored at −20 °C.
The peptides p2-16 and p16-31, corresponding to GCN4p residues 2-16 and 16-31, respectively were synthesized using FMOC solid-phase synthesis. The p2-16 peptide was purchased from Biopeptide (San Diego, CA), while p16-31 was a gift from M. Steinmetz (Paul Scherrer Institut, Switzerland). Sample purity and composition were verified using reversed-phase analytical HPLC and mass spectral analysis.
Concentrations of urea stock solutions were determined from their refractive index 63. Molar extinction coefficients of 1,200 and 1,500 M-1 cm-1 (water, pH 7.0) were used to determine the concentrations of GCN4p monomer and the p16-31 peptide, respectively.
NMR assignments for native and denatured states
Previously published 1H and 15N assignments for native GCN4p 26 were extended to include 13Cα, 13Cβ and 13C’ assignments using 3D HNCACB, HNCO, HCCH-TOCSY, and 13C-edited NOESY-HSQC experiments. For the D state in 6M urea assignments were obtained with 3D HNCACB, HNCA, HN(CO)CA, C(CO)NH, HC(CO)NH, and HNCO experiments as implemented in the Varian Biopack. The p2-16 and p16-31 peptides were assigned as described36.
pH titration measurements
Three sets of samples were used to characterize pKa values. Native GCN4p samples consisted of 2.1 mM 13C/15N-labeled monomer and 150 mM NaCl in 10 mM sodium phosphate buffer. Denatured GCN4p samples had 1.8 mM 13C/15N-labeled monomer concentrations in 6 M urea, 150 mM NaCl, 10 mM phosphate. The p2-16 and p16-31 samples had 3 mM peptide, 150 mM NaCl, in 10 mM phosphate. DSS (2,2-dimethyl-2-silapentane-5-sulfonate, Sigma) was used for an internal chemical shift reference standard. All experiments were done at 25 °C, in water (90% H2O / 10 % D2O) to avoid corrections for the effect of deuterium oxide on the pH electrode.
NMR data were acquired on Varian Inova 500 and 600 MHz spectrometers equipped with cryogenic probes. For each pH point 1D 13C-filtered HSQC experiments were used to follow the titration of the Tyr17 and His18 aromatic ring protons. 2D 1H-13C CT-HSQC experiments selective for -CH2- groups were used to follow the titrations of Asp, Glu, Lys and Arg residues. We also collected a 1D 1H NMR spectrum for referencing on DSS for each pH point, and a 1H-15N HSQC spectrum to follow the shifts of backbone amide protons. Ionizable groups for GCN4p in 6 M urea were primarily characterized using 1H and 15N resonances in 2D 1H-15N HSQC experiments, in contrast to the N state where 2D 1H-13C HSQC experiments were primarily used.
1D 13C-filtered HSQC experiments were acquired with spectral windows of 6200 Hz, digitized into 1024 complex points. 2D 1H-13C CT-HSQC experiments were acquired with 1H × 13C sweep widths of 6200 × 7500 Hz, digitized into 2048 × 200 complex points. 2D 1H-15N HSQC experiments were acquired with 1H × 15N sweep widths of 6200 × 1600 Hz, digitized into 1024 × 100 complex points. The experiments for the D-state protein in 6 M urea were obtained with the same parameters except a smaller 15N spectral width of 1290 Hz. We recorded 1H-13C and 1H-15N HSQC experiments at 33 pH points for native GCN4p. For D-state GCN4 we recorded 38 1H-13C HSQCs, and 16 1H-15N HSQCs. Finally, we collected five 3D HCCH-TOCSY (pH 12.5, 7.4, 5.8, 4.5, 2.5) and three 13C-edited NOESY-HSQC (10.5, 6.9, 2.5) data sets for the N-state protein, to more accurately track the shifts of resonances over the course of the titrations. Titrations for the p2-16 and p16-31 peptides were followed with 1H-13C HSQC and 1H-15N HSQC experiments recorded on samples at natural abundance.
pH adjustments were made using 0.2 to 1 μl aliquots of 1 M HCl and NaOH stock solutions. We used an IQ Scientific -240 meter with a metal PH46-SS probe for pH measurements. A three-point electrode calibration (pH 4, 7, 10) was done for all experiments. Solution pH values were measured before and after each experiment, and typical differences were less than 0.1 pH units. The average of these values was used for analyses. For samples in 6 M urea we needed to correct for the pH offset introduced by the effect of urea on the pH electrode 31,39 using the histidine model compound N(α)-acetyl-L-histidine (Acros Organics). pKa values were measured for the compound in 90% H2O/ 10% D2O (pKa = 7.13) and in a solution of 6 M urea in 90% H2O/ 10% D2O (pKa = 7.67), giving the correction ΔpKa(H2O-urea) = -0.52 pH units.
pKa data analysis
The parameters pKa (ionization constant), δlow (low pH chemical shift plateau), δhigh (high pH chemical shift plateau), and n (Hill coefficient) were determined from least squares fits of the chemical shift (δ) data as a function of pH to the Henderson-Hasselbach equation 27,64:
| (2) |
Together these four parameters can be used to reconstruct chemical shifts for a given resonance at any pH. Titration parameters for native GCN4p, denatured GCN4p, and the 16-31 peptide are given in Supplementary Tables I-III. In cases where multiple resonances could be used to probe the titration of a single residue (e.g. Cγ, Hγ1 and Hγ2 of Glu6), the pKa reported in Table 1 is the average of these values.
Urea unfolding transitions at different pH values
Equilibrium urea unfolding transitions were monitored by circular dichroism (CD) spectroscopy using a Jasco J-810 spectropolarimeter equipped with a 2mm pathlength quartz cuvette. The temperature was maintained at 25 °C with the instrument’s Peltier device. Samples with 30 μM GCN4p were prepared in 25 mM citrate/phosphate/borate buffer, at the appropriate pH. Unfolding was measured from the ellipticity change at 222 nm as a function of urea concentration. Urea solutions were freshly prepared in the appropriate buffer on the day of the experiment.
To obtain thermodynamic parameters for unfolding, the urea concentration dependence of Θ222 was fit to a denaturation equation appropriate for a dimer 65:
| (3) |
Here BN and BU are the baseline CD values for the N and D states, bN and bU are the slopes of the pre- and post-transitions, [P] is the total protein concentration, ΔG0 is the standard state free energy difference in the absence of denaturants, and the m-value is the slope of the unfolding transition. Non-linear fits of Equation 3 were obtained with the program Kaleidagraph 3.6, with the six parameters BN, bN, BU,bU, m, and ΔG0 as free variables in the fit. The ΔG0 and m-values from the fits are given in Supplementary Table IV. Alternatively, we carried out a 5-parameter fit where BN was fixed to the value of the maximum mean residue ellipticity in the absence of denaturant at pH 9, with the assumption that this ellipticity (-31 × 103 deg•cm2•dmol-1) corresponds to 100% native protein. This approach gave similar ΔG0 and m-values as the more standard 6-parameter fit, and the results are presented in Supplementary Table 4. We favor the 6-parameter fit because it makes less assumptions about the ellipticity of the native state.
Structure-based pKa calculations
All calculations were performed with the wild type GCN4p X-ray structure 15 (PDB code 2zta.pdb). The sequence of the protein used in this work is slightly different from that used to determine the crystal structure. Therefore the N-terminal Arg residue of the X-ray structure was replaced with Gly-Ser, and the C-terminal Arg residue was eliminated. After the substitutions were performed in silico, the protein was subjected to 500 steps of steepest descent minimization. pKa values and electrostatic free energies were calculated using methods based on the finite difference solution of the linearized Poisson-Boltzmann equation (FDPB) 66. The UHBD software package was used for this purpose 47,67,68. The full-charge method with the PARSE parameter set 69 was used to model the charge of each ionizable state 68,70,71. The calculations of ionization energies and mean charges were performed using the cluster method 72. In addition to the set of reference pKa values normally used in these calculations 47 the pH dependent properties were also calculated using the set of GCN4p pKa values measured by NMR in urea or from peptide fragments (this work) and with the pKa values measured in random-coil model peptides by potentiometry 34. In the calculations, the internal dielectric constant was set to 20, the dielectric constant of water was 78.5, the water accessible surface of the protein was defined with a probe radius of 1.4 Å 73, the temperature was 298 K, the ionic strength was 150 mM, and the Stern layer - defined as a region inaccessible to counterions was modeled with a probe radius of 2.0 Å 74. The tautomer states used were Asp: OD2, Glu: OE2, His: ND. The method of focusing was implemented 47 which uses an initial lattice size sufficiently large to include the entire molecule and an adequate solvent region at least one-third the longest dimension of the molecule. Previous studies have shown that these calculations can successfully model the ionization energetics of surface ionizable groups 75,76.
Supplementary Material
Acknowledgments
We thank Michel Steinmetz (Paul Scherrer Institut, Switzerland) for the p16-31 peptide. W.M.M. is grateful for a summer graduate research stipend from the Richard C. Crain, Jr. Memorial Fellowship Fund. RAK is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science. BGME’s work was supported by NSF grant MCB 0212414. ATA acknowledges research support from NSF grant MCB 0236316 and funding from NIH-NCRR for NMR instrumentation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bosshard HR, Marti DN, Jelesarov I. Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit. 2004;17:1–16. doi: 10.1002/jmr.657. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein AV, Ptitsyn OB. Protein Physics: A Course of Lectures. Academic Press; Amsterdam: 2002. [Google Scholar]
- 3.Kumar S, Nussinov R. Close-range electrostatic interactions in proteins. Chembiochem. 2002;3:604–17. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Warshel A, Papazyan A. Electrostatic effects in macromolecules: fundamental concepts and practical modeling. Curr Opin Struct Biol. 1998;8:211–7. doi: 10.1016/s0959-440x(98)80041-9. [DOI] [PubMed] [Google Scholar]
- 5.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 6.Sternberg MJ, Hayes FR, Russell AJ, Thomas PG, Fersht AR. Prediction of electrostatic effects of engineering of protein charges. Nature. 1987;330:86–8. doi: 10.1038/330086a0. [DOI] [PubMed] [Google Scholar]
- 7.Sheinerman FB, Honig B. On the role of electrostatic interactions in the design of protein-protein interfaces. J Mol Biol. 2002;318:161–77. doi: 10.1016/S0022-2836(02)00030-X. [DOI] [PubMed] [Google Scholar]
- 8.Ripoll DR, Vila JA, Scheraga HA. On the orientation of the backbone dipoles in native folds. Proc Natl Acad Sci U S A. 2005;102:7559–64. doi: 10.1073/pnas.0502754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones S, Shanahan HP, Berman HM, Thornton JM. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–98. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onufriev A, Case DA, Ullmann GM. A novel view of pH titration in biomolecules. Biochemistry. 2001;40:3413–9. doi: 10.1021/bi002740q. [DOI] [PubMed] [Google Scholar]
- 11.Lavigne P, Sonnichsen FD, Kay CM, Hodges RS. Interhelical salt bridges, coiled-coil stability, and specificity of dimerization. Science. 1996;271:1136–8. doi: 10.1126/science.271.5252.1136. [DOI] [PubMed] [Google Scholar]
- 12.Lumb KJ, Kim PS. Interhelical salt bridges, coiled-coil stability, and specificity of dimerization. Science. 1996;271:1136–8. doi: 10.1126/science.271.5252.1136. [DOI] [PubMed] [Google Scholar]
- 13.Ibarra-Molero B, Zitzewitz JA, Matthews CR. Salt-bridges can stabilize but do not accelerate the folding of the homodimeric coiled-coil peptide GCN4-p1. J Mol Biol. 2004;336:989–96. doi: 10.1016/j.jmb.2003.12.069. [DOI] [PubMed] [Google Scholar]
- 14.Meier M, Burkhard P. Statistical analysis of intrahelical ionic interactions in alpha-helices and coiled coils. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 15.O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–44. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 16.Marqusee S, Baldwin RL. Helix stabilization by Glu-…Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci U S A. 1987;84:8898–902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lumb KJ, Kim PS. Measurement of interhelical electrostatic interactions in the GCN4 leucine zipper. Science. 1995;268:436–9. doi: 10.1126/science.7716550. [DOI] [PubMed] [Google Scholar]
- 18.McLachlan AD, Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 19.Burkhard P, Kammerer RA, Steinmetz MO, Bourenkov GP, Aebi U. The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure Fold Des. 2000;8:223–30. doi: 10.1016/s0969-2126(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 20.Lee DL, Ivaninskii S, Burkhard P, Hodges RS. Unique stabilizing interactions identified in the two-stranded alpha-helical coiled-coil: crystal structure of a cortexillin I/GCN4 hybrid coiled-coil peptide. Protein Sci. 2003;12:1395–405. doi: 10.1110/ps.0241403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–61. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 22.Dames SA, Kammerer RA, Moskau D, Engel J, Alexandrescu AT. Contributions of the ionization states of acidic residues to the stability of the coiled coil domain of matrilin-1. FEBS Lett. 1999;446:75–80. doi: 10.1016/s0014-5793(99)00186-6. [DOI] [PubMed] [Google Scholar]
- 23.Marti DN, Jelesarov I, Bosshard HR. Interhelical ion pairing in coiled coils: solution structure of a heterodimeric leucine zipper and determination of pKa values of Glu side chains. Biochemistry. 2000;39:12804–18. doi: 10.1021/bi001242e. [DOI] [PubMed] [Google Scholar]
- 24.Marti DN, Bosshard HR. Electrostatic interactions in leucine zippers: thermodynamic analysis of the contributions of Glu and His residues and the effect of mutating salt bridges. J Mol Biol. 2003;330:621–37. doi: 10.1016/s0022-2836(03)00623-5. [DOI] [PubMed] [Google Scholar]
- 25.Hendsch ZS, Tidor B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 1994;3:211–26. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kammerer RA, Jaravine VA, Frank S, Schulthess T, Landwehr R, Lustig A, Garcia-Echeverria C, Alexandrescu AT, Engel J, Steinmetz MO. An intrahelical salt bridge within the trigger site stabilizes the GCN4 leucine zipper. J Biol Chem. 2001;276:13685–8. doi: 10.1074/jbc.M010492200. [DOI] [PubMed] [Google Scholar]
- 27.Markley JL. Observation of histidine residues in proteins by means of nuclear magnetic resonance spectroscopy. Accounts of Chemical Research. 1975;8:70–80. [Google Scholar]
- 28.Tollinger M, Crowhurst KA, Kay LE, Forman-Kay JD. Site-specific contributions to the pH dependence of protein stability. Proc Natl Acad Sci U S A. 2003;100:4545–50. doi: 10.1073/pnas.0736600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan YJ, Oliveberg M, Davis B, Fersht AR. Perturbed pKa-values in the denatured states of proteins. J Mol Biol. 1995;254:980–92. doi: 10.1006/jmbi.1995.0670. [DOI] [PubMed] [Google Scholar]
- 30.Kuhlman B, Luisi DL, Young P, Raleigh DP. pKa values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry. 1999;38:4896–903. doi: 10.1021/bi982931h. [DOI] [PubMed] [Google Scholar]
- 31.Anderson DE, Becktel WJ, Dahlquist FW. pH-induced denaturation of proteins: a single salt bridge contributes 3-5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry. 1990;29:2403–8. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- 32.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy: principles and practice. Academic Press; San Diego: 1996. [Google Scholar]
- 33.Yamazaki T, Nicholson LK, Torchia DA, Wingfield P, Stahl SJ, Kaufman JD, Eyermann CJ, Hodge CN, Lam PYS, Ru Y, Jadhav PK, Chang C-H, Weber PC. NMR and X-Ray Evidence that the HIV Protease Catalytic Aspartyl Groups Are Protonated in the Complex Formed by the Protease and a Non-Peptide Cyclic Urea-Based Inhibitor. J Am Chem Soc. 1994;116:10791–10792. [Google Scholar]
- 34.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein Sci. 2006;15:1214–8. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrescu AT, Evans PA, Pitkeathly M, Baum J, Dobson CM. Structure and dynamics of the acid-denatured molten globule state of alpha-lactalbumin: a two-dimensional NMR study. Biochemistry. 1993;32:1707–18. doi: 10.1021/bi00058a003. [DOI] [PubMed] [Google Scholar]
- 36.Steinmetz MO, Jelesarov I, Matousek WM, Honnappa S, Jahnke W, Missimer JH, Frank S, Alexandrescu AT, Kammerer RA. Molecular basis of coiled-coil formation. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0700321104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kammerer RA, Schulthess T, Landwehr R, Lustig A, Engel J, Aebi U, Steinmetz MO. An autonomous folding unit mediates the assembly of two-stranded coiled coils. Proc Natl Acad Sci U S A. 1998;95:13419–24. doi: 10.1073/pnas.95.23.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pujato M, Navarro A, Versace R, Mancusso R, Ghose R, Tasayco ML. The pH-dependence of amide chemical shift of Asp/Glu reflects its pKa in intrinsically disordered proteins with only local interactions. Biochim Biophys Acta. 2006;1764:1227–33. doi: 10.1016/j.bbapap.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Marti DN. Apparent pKa shifts of titratable residues at high denaturant concentration and the impact on protein stability. Biophys Chem. 2005;118:88–92. doi: 10.1016/j.bpc.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Mira MM, Sanchez-Ruiz JM. pH Corrections and Protein Ionization in Water/Guanidinium Chloride. Biophysical J. 2001;81:3489–3502. doi: 10.1016/S0006-3495(01)75980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acevedo O, Guzman-Casado M, Garcia-Mira MM, Ibarra-Molero B, Sanchez-Ruiz JM. pH Corrections in Chemical Denaturant Solutions. Analytical Biochemistry. 2002;306:158–161. doi: 10.1006/abio.2002.5668. [DOI] [PubMed] [Google Scholar]
- 42.Nikolaev Y, Pervushin K. NMR spin state exchange spectroscopy reveals equilibrium of two distinct conformations of leucine zipper GCN4 in solution. J Am Chem Soc. 2007;129:6461–6469. doi: 10.1021/ja0685295. [DOI] [PubMed] [Google Scholar]
- 43.Whitten ST, Garcia-Moreno EB. pH dependence of stability of staphylococcal nuclease: evidence of substantial electrostatic interactions in the denatured state. Biochemistry. 2000;39:14292–304. doi: 10.1021/bi001015c. [DOI] [PubMed] [Google Scholar]
- 44.Zhou HX. A Gaussian-chain model for treating residual charge-charge interactions in the unfolded state of proteins. Proc Natl Acad Sci U S A. 2002;99:3569–74. doi: 10.1073/pnas.052030599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou HX. Direct test of the Gaussian-chain model for treating residual charge-charge interactions in the unfolded state of proteins. J Am Chem Soc. 2003;125:2060–1. doi: 10.1021/ja0298491. [DOI] [PubMed] [Google Scholar]
- 46.Fitch CA, Whitten ST, Hilser VJ, Garcia-Moreno EB. Molecular mechanisms of pH-driven conformational transitions of proteins: insights from continuum electrostatics calculations of acid unfolding. Proteins. 2006;63:113–26. doi: 10.1002/prot.20797. [DOI] [PubMed] [Google Scholar]
- 47.Antosiewicz J, McCammon JA, Gilson MK. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994;238:415–36. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharya S, Lecomte JT. Temperature dependence of histidine ionization constants in myoglobin. Biophys J. 1997;73:3241–56. doi: 10.1016/S0006-3495(97)78349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halle B. Biomolecular cryocrystallography: structural changes during flash-cooling. Proc Natl Acad Sci U S A. 2004;101:4793–8. doi: 10.1073/pnas.0308315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antosiewicz J, McCammon JA, Gilson MK. The determinants of pKas in proteins. Biochemistry. 1996;35:7819–33. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]
- 51.Kim J, Mao J, Gunner MR. Are acidic and basic groups in buried proteins predicted to be ionized? J Mol Biol. 2005;348:1283–98. doi: 10.1016/j.jmb.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 52.Linse S, Jonsson B, Chazin WJ. The effect of protein concentration on ion binding. Proc Natl Acad Sci U S A. 1995;92:4748–52. doi: 10.1073/pnas.92.11.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker WR, Kintanar A. Characterization of the pH titration shifts of ribonuclease A by one- and two-dimensional nuclear magnetic resonance spectroscopy. Arch Biochem Biophys. 1996;327:189–99. doi: 10.1006/abbi.1996.0108. [DOI] [PubMed] [Google Scholar]
- 54.Berisio R, Lamzin VS, Sica F, Wilson KS, Zagari A, Mazzarella L. Protein titration in the crystal state. J Mol Biol. 1999;292:845–54. doi: 10.1006/jmbi.1999.3093. [DOI] [PubMed] [Google Scholar]
- 55.Barlow DJ, Thornton JM. Ion-pairs in proteins. J Mol Biol. 1983;168:867–85. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 56.Khare D, Alexander P, Antosiewicz J, Bryan P, Gilson M, Orban J. pKa measurements from nuclear magnetic resonance for the B1 and B2 immunoglobulin G-binding domains of protein G: comparison with calculated values for nuclear magnetic resonance and X-ray structures. Biochemistry. 1997;36:3580–9. doi: 10.1021/bi9630927. [DOI] [PubMed] [Google Scholar]
- 57.Makhatadze GI, Loladze VV, Ermolenko DN, Chen X, Thomas ST. Contribution of surface salt bridges to protein stability: guidelines for protein engineering. J Mol Biol. 2003;327:1135–48. doi: 10.1016/s0022-2836(03)00233-x. [DOI] [PubMed] [Google Scholar]
- 58.van Vlijmen HW, Schaefer M, Karplus M. Improving the accuracy of protein pKa calculations: conformational averaging versus the average structure. Proteins. 1998;33:145–58. doi: 10.1002/(sici)1097-0134(19981101)33:2<145::aid-prot1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 59.Zhou HX, Vijayakumar M. Modeling of protein conformational fluctuations in pKa predictions. J Mol Biol. 1997;267:1002–11. doi: 10.1006/jmbi.1997.0895. [DOI] [PubMed] [Google Scholar]
- 60.You TJ, Bashford D. Conformation and hydrogen ion titration of proteins: a continuum electrostatic model with conformational flexibility. Biophys J. 1995;69:1721–33. doi: 10.1016/S0006-3495(95)80042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kammerer RA, Schulthess T, Landwehr R, Lustig A, Fischer D, Engel J. Tenascin-C hexabrachion assembly is a sequential two-step process initiated by coiled-coil alpha-helices. J Biol Chem. 1998;273:10602–8. doi: 10.1074/jbc.273.17.10602. [DOI] [PubMed] [Google Scholar]
- 62.Wiltscheck R, Kammerer RA, Dames SA, Schulthess T, Blommers MJ, Engel J, Alexandrescu AT. Heteronuclear NMR assignments and secondary structure of the coiled coil trimerization domain from cartilage matrix protein in oxidized and reduced forms. Protein Sci. 1997;6:1734–45. doi: 10.1002/pro.5560060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–80. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 64.Dames SA, Kammerer RA, Wiltscheck R, Engel J, Alexandrescu AT. NMR structure of a parallel homotrimeric coiled coil. Nat Struct Biol. 1998;5:687–91. doi: 10.1038/90444. [DOI] [PubMed] [Google Scholar]
- 65.Zitzewitz JA, Bilsel O, Luo J, Jones BE, Matthews CR. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry. 1995;34:12812–9. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 66.Warwicker J, Watson HC. Calculation of the electric potential in the active site cleft due to alpha-helix dipoles. J Mol Biol. 1982;157:671–9. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- 67.Davis ME, Madura JD, Luty BA, McCammon JA. Electrostatics and diffusion of molecules in solution - simulation with the University of Houston Brownian Dynamics program. Computer Physics Communications. 1991;62:187–191. [Google Scholar]
- 68.Antosiewicz J, Briggs JM, Elcock AH, Gilson MK, McCammon JA. Computing ionization states of proteins with a detailed charge model. Journal of Computational Chemistry. 1996;17:1633–1644. [Google Scholar]
- 69.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. Journal of Physical Chemistry. 1994;98:1978–1988. [Google Scholar]
- 70.Yang AS, Gunner MR, Sampogna R, Sharp K, Honig B. On the calculation of pKas in proteins. Proteins. 1993;15:252–65. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- 71.Bashford D, Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992;224:473–86. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 72.Gilson MK. Multiple-site titration and molecular modeling: two rapid methods for computing energies and forces for ionizable groups in proteins. Proteins. 1993;15:266–82. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- 73.Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–76. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 74.Gilson MK, Honig BH. Energetics of charge-charge interactions in proteins. Proteins. 1988;3:32–52. doi: 10.1002/prot.340030104. [DOI] [PubMed] [Google Scholar]
- 75.Lee KK, Fitch CA, Garcia-Moreno EB. Distance dependence and salt sensitivity of pairwise, coulombic interactions in a protein. Protein Sci. 2002;11:1004–16. doi: 10.1110/ps.4700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee KK, Fitch CA, Lecomte JT, Garcia-Moreno EB. Electrostatic effects in highly charged proteins: salt sensitivity of pKa values of histidines in staphylococcal nuclease. Biochemistry. 2002;41:5656–67. doi: 10.1021/bi0119417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


