Abstract
Biosynthesis of the Salmonella typhimurium LT2 O antigen is encoded by genes which map in the rfb cluster. The cloning and restriction enzyme analysis of part of this cluster have been described previously (H. N. Brahmbhatt, N. B. Quigley, and P. R. Reeves, Mol. Gen. Genet. 203:172-176, 1986). The entire rfb gene cluster has now been cloned, and a detailed restriction enzyme map has been constructed which has enabled us to map the approximate positions of individual rfb genes.
Full text
PDF
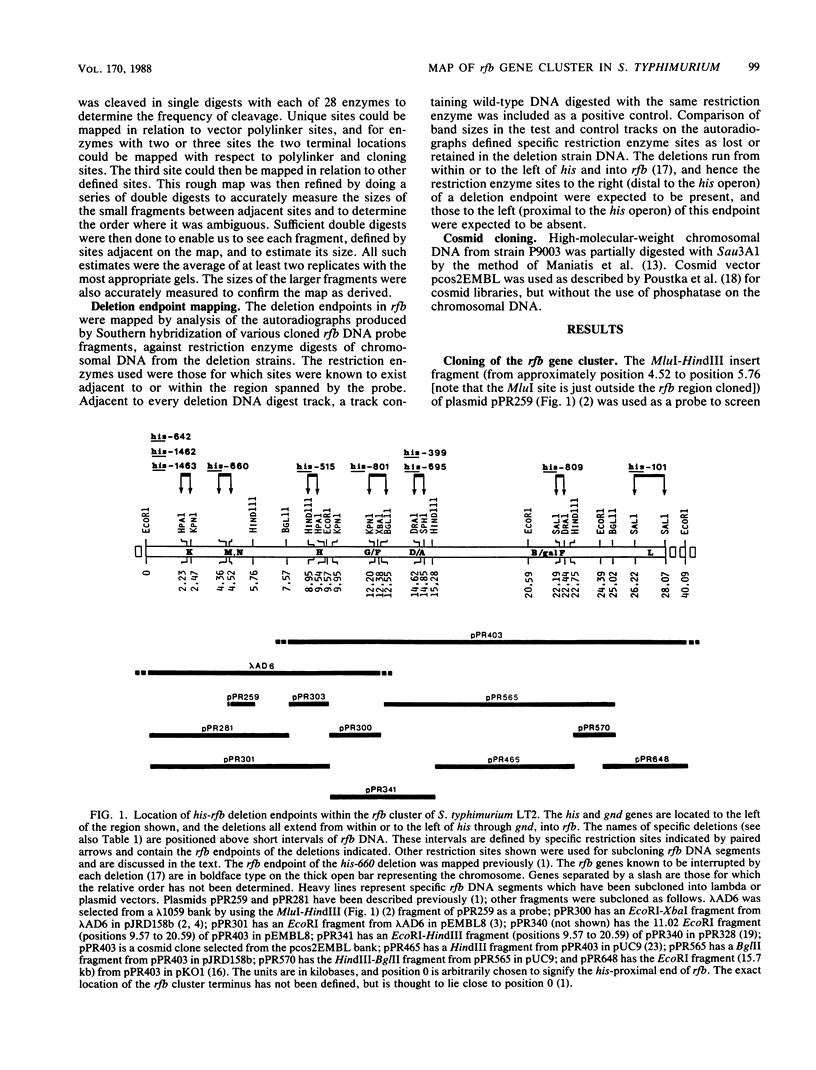


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brahmbhatt H. N., Quigley N. B., Reeves P. R. Cloning part of the region encoding biosynthetic enzymes for surface antigen (O-antigen) of Salmonella typhimurium. Mol Gen Genet. 1986 Apr;203(1):172–176. doi: 10.1007/BF00330399. [DOI] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Merchez M., Brunel F. Vectors with restriction-site banks. I. pJRD158, a 3903-bp plasmid containing 28 unique cloning sites. Gene. 1984 Jun;28(3):311–318. doi: 10.1016/0378-1119(84)90148-3. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusterspreute M., Davison J. Restriction site bank vectors. II. DNA sequence analysis of plasmid pJRD158. DNA. 1984 Jun;3(3):259–268. doi: 10.1089/dna.1.1984.3.259. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal M., Nikaido H. Consequences of deletion mutations joining two operons of opposite polarity. J Mol Biol. 1969 Jun 28;42(3):511–520. doi: 10.1016/0022-2836(69)90239-3. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Maurer R., Osmond B. C., Shekhtman E., Wong A., Botstein D. Functional interchangeability of DNA replication genes in Salmonella typhimurium and Escherichia coli demonstrated by a general complementation procedure. Genetics. 1984 Sep;108(1):1–23. doi: 10.1093/genetics/108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Nikaido H., Levinthal M., Nikaido K., Nakane K. Extended deletions in the histidine-rough-B region of the Salmonella chromosome. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1825–1832. doi: 10.1073/pnas.57.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka A., Rackwitz H. R., Frischauf A. M., Hohn B., Lehrach H. Selective isolation of cosmid clones by homologous recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4129–4133. doi: 10.1073/pnas.81.13.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley N. B., Reeves P. R. Chloramphenicol resistance cloning vector based on pUC9. Plasmid. 1987 Jan;17(1):54–57. doi: 10.1016/0147-619x(87)90008-4. [DOI] [PubMed] [Google Scholar]
- Ratcliff S. W., Luh J., Ganesan A. T., Behrens B., Thompson R., Montenegro M. A., Morelli G., Trautner T. A. The genome of Bacillus subtilis phage SPP1: the arrangement of restriction endonuclease generated fragments. Mol Gen Genet. 1979 Jan 10;168(2):165–172. doi: 10.1007/BF00431442. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, Edition VI. Microbiol Rev. 1983 Sep;47(3):410–453. doi: 10.1128/mr.47.3.410-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N. K., Quigley N. B., Reeves P. R. O-antigen variation in Salmonella spp.: rfb gene clusters of three strains. J Bacteriol. 1988 Jan;170(1):103–107. doi: 10.1128/jb.170.1.103-107.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yuasa R., Levinthal M., Nikaido H. Biosynthesis of cell wall lipopolysaccharide in mutants of Salmonella. V. A mutant of Salmonella typhimurium defective in the synthesis of cytidine diphosphoabequose. J Bacteriol. 1969 Oct;100(1):433–444. doi: 10.1128/jb.100.1.433-444.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]