Abstract
The chemokine receptor CXCR4 functions as a fusion coreceptor for T cell tropic and dual-tropic HIV-1 strains. To identify regions of CXCR4 that are important for coreceptor function, CXCR4–CXCR2 receptor chimeras were tested for the ability to support HIV-1 envelope (env) protein-mediated membrane fusion. Receptor chimeras containing the first and second extracellular loops of CXCR4 supported fusion by T tropic and dual-tropic HIV-1 and HIV-2 strains and binding of a monoclonal antibody to CXCR4, 12G5, that blocks CXCR4-dependent infection by some virus strains. The second extracellular loop of CXCR4 was sufficient to confer coreceptor function to CXCR2 for most virus strains tested but did not support binding of 12G5. Truncation of the CXCR4 cytoplasmic tail or mutation of a conserved DRY motif in the second intracellular loop did not affect coreceptor function, indicating that phosphorylation of the cytoplasmic tail and the DRY motif are not required for coreceptor function. The results implicate the involvement of multiple CXCR4 domains in HIV-1 coreceptor function, especially the second extracellular loop, though the structural requirements for coreceptor function were somewhat variable for different env proteins. Finally, a hybrid receptor in which the amino terminus of CXCR4 was replaced by that of CCR5 was active as a coreceptor for M tropic, T tropic, and dual-tropic env proteins. We propose that dual tropism may evolve in CCR5-restricted HIV-1 strains through acquisition of the ability to utilize the first and second extracellular loops of CXCR4 while retaining the ability to interact with the CCR5 amino-terminal domain.
The envelope (env) protein of HIV-1 binds to CD4 with high affinity and mediates fusion between the viral and cellular membranes (for review, see ref. 1). The membrane fusion reaction results from conformational changes in env that likely include the formation of a coiled-coil structure with the resulting exposure of the amino-terminal fusion peptide in gp41 (2, 3). Although binding to CD4 clearly triggers conformational changes in env, including increased exposure of the V3 loop, these changes by themselves are not sufficient for the membrane fusion reaction (4). Evidence for this comes from observations that expression of CD4 does not render most nonhuman cells susceptible to env-mediated membrane fusion and virus entry (5–8). In addition, HIV-1 strains exhibit distinct tropisms for CD4-positive cells. Macrophage tropic (M tropic) strains enter and replicate in macrophages and primary T cells but generally fail to enter T cell lines, whereas T cell tropic strains fail to enter macrophages efficiently (for review, see ref. 9). Dual-tropic viruses, which may represent an intermediate form during the evolution from M to T tropism, can enter all three target cell types. These findings indicated that CD4 and one or more cellular cofactors or coreceptors are required for membrane fusion and infection to occur and that these coreceptors may play an important role in determining viral tropism.
The inability of CD4 to render nonhuman cells permissive for env-mediated syncytia formation was used to identify cDNAs that could, in conjunction with CD4, make murine cells permissive for cell–cell fusion by the T cell tropic BH8 env glycoprotein. The cDNA that imparted this phenotype was found to encode an orphan seven-transmembrane-domain receptor that supported membrane fusion by T cell tropic, but not M tropic, HIV-1 env proteins in a CD4-dependent fashion (10). This protein, termed fusin, was designated CXCR4 when it was shown to bind the CXC chemokine SDF-1 (11, 12). The discovery of fusin coupled with the finding that the chemokines RANTES (regulated upon activation, normal T cell expressed and secreted), MIP-1α, and MIP-1β (where MIP is macrophage inflammatory protein) could block infection by M tropic virus strains (13) rapidly led to the discovery that CCR5 was the fusion coreceptor for these strains (14–18). The importance of chemokine receptors for virus infection in vivo was shown by the discovery of a CCR5 polymorphism for which approximately 1% of the Caucasian population is homozygous (19, 20). Cells from these individuals are highly resistant to infection by M tropic CCR5-restricted viruses both in vitro and in vivo (19–22).
Given the critical role of chemokine receptors in virus entry and the ability of chemokines to block virus infection, it will be important to elucidate the mechanism by which they allow membrane fusion to occur. Recently, the gp120 subunit of a T tropic env was shown to interact directly with CXCR4 (23), whereas M tropic env proteins interact with CCR5 in a CD4-dependent fashion (24, 25). Although much progress has been made in identifying regions of CCR5 required for HIV-1 and simian immunodeficiency virus coreceptor function (27–29), little is known about the structure–function relationships of CXCR4. To identify the structural determinants required for CXCR4 coreceptor function, we constructed chimeric receptors based on CXCR4 and CXCR2, a chemokine receptor that lacks HIV-1 coreceptor activity. The chimeric receptors were studied for their ability to support env-mediated membrane fusion and to bind the conformation-dependent anti-CXCR4 monoclonal antibody 12G5 (30). We found that there is differential CXCR4 utilization by diverse env proteins, that the second extracellular loop of CXCR4 was sufficient for coreceptor activity for most virus strains tested, and that the epitope for 12G5 is contained in extracellular loops 1 and 2. Our results suggest that M tropic CCR5-restricted viruses can evolve to become dual tropic by acquiring the ability to interact with the extracellular loops of CXCR4 while retaining the ability to productively interact with the CCR5 amino-terminal domain.
MATERIALS AND METHODS
Constructs.
Chimeric receptors composed of CXCR4 and CXCR2 or CCR5 segments were constructed by the PCR–ligation–PCR approach. To create chimeras, complementary regions of the two parental receptors were amplified from cDNA templates, and the blunt-ended amplification products were ligated after phosphorylation. The product encoding the desired hybrid was then amplified with the appropriate upstream and downstream primers from the respective parental receptors. Products of the predicted size were cloned into the TA vector (Invitrogen), screened, and sequenced. The final cDNA encoding the chimeric receptor was subcloned into the pcDNA3 expression vector (Invitrogen) by using unique sites introduced into the PCR primers. Primer sequences can be obtained from the authors. The plasmid luciferase-T7, encoding firefly luciferase under control of the T7 promoter, was obtained from Promega. pT4 encodes CD4 under control of the cytomegalovirus promoter. The rat CXCR4 homolog was provided by Richard Duman (Yale University) and was cloned into pSP73.
Cells and Viruses.
HeLa and QT6 cells were cultured as described (31). A panel of recombinant vaccinia viruses encoding HIV env proteins were used. These included vSC60 (BH8) (32), vCB28 (JR-FL) (32), vCB36 (RF) (32), vBD3 (89.6) (17), vSC50 (HIV-2SBL6669) (33), and vCB51 [BK132, a primary T tropic clade B virus (34)]. The recombinant vaccinia virus vTF1.1, which encodes T7 RNA polymerase, was also used (35).
Gene Reporter Fusion Assay.
A previously described luciferase-based gene reporter assay was used to quantitate cell–cell fusion events (17, 29, 36). Briefly, T7 RNA polymerase and HIV-1 envelope proteins were introduced into effector HeLa cells by recombinant vaccinia viruses. Luciferase-T7, CD4, and coreceptors were introduced into target QT6 cells by calcium phosphate-mediated transfection. After overnight incubation, effector cells were added to the target cells to initiate fusion. Seven to 8 h after mixing, the cells were lysed and assayed for luciferase activity.
Flow Cytometry Analysis.
QT6 cells were transiently transfected with wild-type and chimeric cofactors by calcium phosphate precipitation. After expression for 16–20 h, cells were removed from the plate with 1 mM EDTA, centrifuged, and resuspended in PBS with bovine serum albumin (1 g/liter) supplemented with 1% normal rat sera and 1% normal rabbit sera and placed on ice. Chimeras containing the amino terminus of CXCR2 were stained with the 10G2 monoclonal antibody (37), while those containing the amino terminus of CXCR4 were stained with rabbit polyvalent antibodies produced by immunization with glutathione S-transferase fusion proteins containing this domain of CXCR4. Cells were also stained with 12G5, a monoclonal antibody to CXCR4, to map its binding site (30).
RESULTS
Replacement of Single CXCR4 Domains with Corresponding CXCR2 Regions.
Introduction of CXCR4 into CD4-positive nonpermissive cells makes them susceptible to T tropic and dual-tropic HIV-1 infection and envelope-mediated cell–cell fusion (10, 15–18, 31, 38). To identify regions of CXCR4 required for coreceptor function, we constructed chimeric molecules between CXCR4 and CXCR2 (Fig. 1), which share approximately 35% amino acid identity. However, CXCR2 has not been reported to serve as a HIV-1 coreceptor. The more closely related rat CXCR4 homolog [86% identity (39)] functioned as an HIV-1 fusion coreceptor for HIV-1 BH8 and, therefore, could not be used for the construction of CXCR4 chimeras (data not shown). CXCR2 was also chosen because of the availability of a monoclonal antibody to the amino-terminal domain (10G2) (37). This antibody, along with a monoclonal antibody to CXCR4 (12G5) (30) and a rabbit polyclonal serum to the CXCR4 amino terminus, made it possible to monitor surface expression of the chimeric molecules. However, because of the necessity to use three antibodies, of the diverse structures of the chimeric receptors that might alter antibody affinity, and the chimeric receptors were expressed transiently, precise comparisons of surface expression levels were not possible. Therefore, greater emphasis should be placed on chimeric receptors that allow cell–cell fusion rather than those that do not.
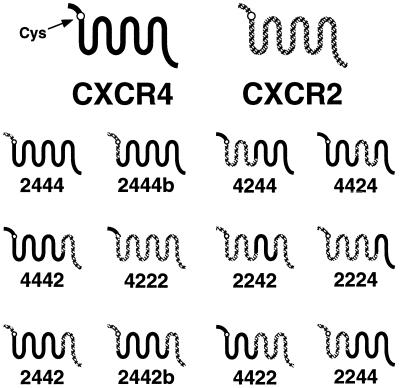
Figure 1.
CXCR4–CXCR2 chimeras. Chimeric receptors based on CXCR4 and CXCR2 are depicted schematically. Amino-terminal domain exchanges were performed at the conserved cysteine residue at position 28 of CXCR4.
The ability of chimeric receptors to serve as coreceptors was determined with a cell–cell fusion assay in which effector cells that express HIV env are mixed with target cells that express CD4 and a candidate coreceptor (17, 29). In addition, effector cells are infected with a recombinant vaccinia virus that expresses T7 RNA polymerase, and target cells are transfected with a plasmid encoding luciferase under the transcriptional control of the T7 promoter. Although HeLa cells were used as effectors, the QT6 quail fibrosarcoma cell line served as targets because they do not support HIV env-mediated membrane fusion when they express CD4 alone, and they can be transfected at high efficiency. The target and effector cells are mixed at 37°C, and if fusion occurs, cytoplasmic mixing between effectors and targets results in expression of luciferase, which can be easily measured. Thus far, we have found perfect concordance between the ability of wild-type or mutant chemokine receptors to support cell fusion and their ability to support virus entry (17, 20, 27, 29, 31).
To determine whether any single CXCR4 domain is required for coreceptor activity, we substituted single CXCR4 domains with corresponding regions from CXCR2 (Fig. 1). The parental molecule from which each extracellular domain was derived is indicated by the name of each chimera. Thus, chimera 2444 contains the amino-terminal domain of CXCR2 (up to the conserved cysteine residue) in a CXCR4 background. We measured the ability of the chimeric receptors to support cell–cell fusion by T tropic (BH8 and BK132) and dual-tropic (89.6 and RF) HIV-1 env proteins and by HIV-2SBL6669. Although HIV-1 RF is a T tropic virus, we have found that the RF env protein is dual tropic in that it can use both CXCR4 and CCR5 as coreceptors for cell–cell fusion (see Fig. 4). This panel of proteins was chosen because we have shown that env proteins can interact with CCR5 differently (27, 29). Thus, by examining several T and dual-tropic env proteins, we sought to identify differences in structural requirements for the coreceptor activity of CXCR4 by different env glycoproteins and to minimize the possibility of falsely scoring a chimeric receptor as negative for coreceptor activity when examining only a single env protein.
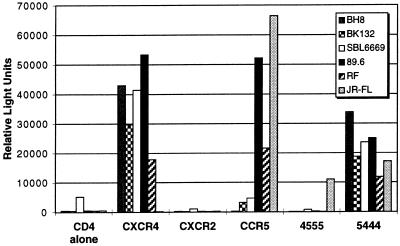
Figure 4.
CXCR4–CCR5 chimeras. A receptor chimera containing the amino-terminal domain of CCR5 (up to the cysteine residue) in a CXCR4 background (5444) and its reciprocal (4555) were tested for coreceptor function in the cell–cell fusion assay described in Fig. 2. Results are expressed in relative light units.
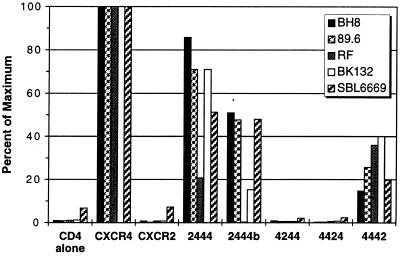
As shown in Fig. 2, fusion was not observed when target cells expressed CD4 alone or CXCR2 and CD4 together but was readily detected when CXCR4 and CD4 were coexpressed. Four of the five env proteins gave similar fusion profiles with the panel of chimeric receptors tested. The BH8, BK132, 89.6, and HIV-2SBL6669 env proteins fused with cells expressing chimeras 2444, 2444b, and 4442 but did not fuse with cells expressing 4244 or 4424. These results indicated that the amino-terminal domain and third extracellular loop of CXCR4 could be individually replaced by the corresponding highly divergent domains of CXCR2 without loss of coreceptor function. However, fusion with cells expressing 2444, 2444b, and 4442 was less efficient than that observed with wild-type CXCR4, suggesting that either the amino-terminal domain and/or the third extracellular loop of CXCR4 might contribute to the fusion reaction, either directly or indirectly. A role for the amino-terminal domain of CXCR4 in coreceptor function was also suggested by our findings with the dual-tropic env protein RF that was particularly sensitive to amino-terminal domain alterations. Replacement of the amino-terminal domain of CXCR4 up to the conserved cysteine residue at position 28 (chimera 2444) resulted in an 80% reduction in fusion activity, whereas replacement of the entire amino-terminal domain (chimera 2444b) resulted in the complete loss of fusion activity.
Figure 2.
Effects of single CXCR2 domains in CXCR4 on coreceptor function. Receptor chimeras composed of single CXCR2 domains in a CXCR4 background were tested for function. HeLa cells expressing the BH8, 89.6, BK132, RF, or HIV-2SBL6669 env proteins in conjunction with T7 RNA polymerase as a consequence of infection with the appropriate recombinant vaccinia virus vectors were mixed with quail QT6 cells transfected with plasmids expressing CD4, the indicated chemokine receptor, and luciferase under control of the T7 promoter. After 8 h at 37°C, the cells were lysed, and the amount of luciferase activity was measured in relative light units and expressed relative to wild-type CXCR4. The signal/noise values for wild-type CXCR4 were typically greater than 50:1.
To determine whether the failure of chimeras 4244 or 4424 to support fusion or the reduced fusion seen with chimeras 2444, 2444b, and 4442 could be accounted for by reduced expression on the cell surface, QT6 cells expressing these molecules were subjected to flow cytometry analysis with a monoclonal antibody to the amino-terminal domain of CXCR2 (10G2) or a polyclonal antibody to the CXCR4 amino terminus. We found that chimeras 2444, 2444b, and 4442 were expressed on the cell surface at levels similar to that seen with CXCR4, but chimeras 4244 and 4424 were not detected (Table 1).
Table 1.
Surface expression and coreceptor function of CXCR4 mutants and chimeras
| Construct | Surface staining | Cell–cell fusion
|
||||
|---|---|---|---|---|---|---|
| BH8 | 89.6 | BK132 | RF | SBL6669 | ||
| CXCR4 | +*† | +++ | +++ | +++ | +++ | +++ |
| CXCR2 | +‡ | − | − | − | − | − |
| 2444 | +*‡ | +++ | +++ | +++ | + | ++ |
| 2444b | +‡ | ++ | ++ | + | − | ++ |
| 4244 | − | − | − | − | − | − |
| 4424 | − | − | − | − | − | − |
| 4442 | +*† | + | + | ++ | ++ | + |
| 4222 | +† | − | − | − | − | − |
| 2242 | +‡ | + | + | ± | − | ND |
| 2224 | − | − | − | − | − | − |
| 2442 | +*‡ | + | + | ± | − | ND |
| 2442b | − | − | − | − | − | − |
| 4422 | − | − | − | − | − | − |
| 2244 | +‡ | − | + | − | − | + |
| CXCR4-tail | +*† | +++ | +++ | ND | ND | ND |
| CXCR4-NAA | +* | + | + | ND | + | ND |
Surface expression after transient expression in QT6 cells was determined by flow cytometry staining with 12G5, rabbit serum to the CXCR4 amino terminus, or 10G2. Fusion activity relative to wild-type CXCR4 was determined as in Figs. 2, 3, 4, with 66–100% = +++; 33–65% = ++; 10–33% = +; 5–10% = ±; 0–5% = −. Fusion results for coreceptors that functioned were the average of at least three experiments. ND, not determined.
12G5 stained cells expressing the indicated receptor.
Rabbit serum to the CXCR4 amino terminus stained cells expressing the indicated receptor.
10G2 stained cells expressing the indicated receptor.
Replacement of Multiple CXCR4 Domains with Corresponding CXCR2 Regions.
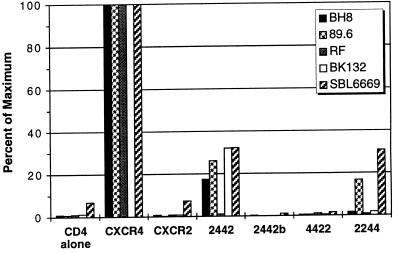
The results in Fig. 2 indicated that both the amino-terminal domain and third extracellular loop of CXCR4 could be individually replaced with the corresponding CXCR2 domains without ablating coreceptor function. To determine whether replacing both of these regions in one molecule could be tolerated, we constructed chimeras in which part or all of the amino terminus and the third extracellular loop of CXCR4 were replaced with the corresponding CXCR2 regions (Fig. 1). Chimera 2442 supported cell–cell fusion for all envs except RF, indicating that the region of CXCR4 from the first cysteine residue through the extracellular loop was sufficient for coreceptor function for most env proteins (Fig. 3). The inability of chimera 2442 to function as a coreceptor for RF indicates that although replacement of the first 27 amino acids of CXCR4 creates a less efficient but functional coreceptor (chimera 2444), this substitution is not tolerated in the presence of a divergent third extracellular loop. As the cysteine residues in the amino-terminal domain and third extracellular loop of CXCR4 are predicted to form a disulfide bond, it is likely that portions of these two domains must interact closely with each other. Chimera 2442b, in which the entire amino-terminal domain of CXCR4 was replaced with that from CXCR2, was not expressed on the cell surface (Table 1).
Figure 3.
Effects of multiple domain substitutions on coreceptor function. The ability of receptor chimeras containing multiple domain substitutions between CXCR4 and CXCR2 to support env-mediated cell–cell fusion by the BH8, 89.6, BK132, RF, or HIV-2SBL6669 env proteins was determined with the assay described in Fig. 2.
To examine the roles of the first and second extracellular loops in coreceptor function more closely, we constructed chimeras 4422 and 2244 (Fig. 1). Chimera 4422 did not support membrane fusion with any of the env glycoproteins; however, this hybrid was expressed on the cell surface at low levels. By contrast, expression of chimera 2244 in conjunction with CD4 supported fusion by the 89.6 and HIV-2SBL6669 env proteins but not by BH8, BK132, or RF env. This observation, in conjunction with the fact that chimera 2442 supported fusion by 89.6 and HIV-2SBL6669, indicates that the second extracellular loop of CXCR4 is critical for coreceptor utilization by these envs. Since BH8 and BK132 can use 2442 but not 2244, it is clear that the first extracellular loop also plays an important role in coreceptor function for these env proteins.
Replacement of Single CXCR2 Domains with Corresponding CXCR4 Regions.
We have found that the amino-terminal domain of CCR5 is sufficient to confer coreceptor activity to CCR2b, CCR1, and CXCR2 (refs. 27 and 29; unpublished data). To determine whether any single extracellular domain of CXCR4 can also confer coreceptor function to heterologous receptors, we constructed receptor chimeras in which single domains of CXCR4 were introduced into CXCR2 (Fig. 1). We found that chimeras 4222 and 2224 failed to support membrane fusion by any of the env proteins tested (Table 1). Flow cytometry analysis indicated that chimera 2224 was not expressed on the cell surface (Table 1). However, since the reciprocal chimera 4442 supports membrane fusion, we consider it unlikely that the third extracellular loop of CXCR4 alone can confer coreceptor function to CXCR2. Cell surface expression of chimera 4222 was detected with polyvalent rabbit antibodies raised against a glutathione S-transferase fusion protein containing the amino-terminal ectodomain of CXCR4. Therefore, the amino-terminal domain of CXCR4 is not sufficient to confer coreceptor activity to CXCR2. Chimera 2242 supported fusion by the BH8, 89.6, and BK132 env proteins, indicating that the second extracellular loop of CXCR4 is sufficient for coreceptor function when placed into CXCR2. Fusion was relatively inefficient, however, suggesting that other CXCR4 domains are also important for full coreceptor function. The ability of 12G5, a conformation-dependent monoclonal antibody to CXCR4, to recognize 2442 but not 2244 or 2242 (Table 1) indicates that the first extracellular loop is a critical part of the 12G5 epitope.
Evolution of Coreceptor Usage Through Distinct CCR5 and CXCR4 Domains.
Our findings that the second extracellular loop of CXCR4 is critical for coreceptor function are in marked contrast to CCR5, where the first 20 residues of CCR5 are sufficient to confer coreceptor function to diverse chemokine receptors, including CCR2b, CCR1, and CXCR2 (ref. 29; unpublished data). However, for most M tropic HIV-1 strains the amino-terminal domain of CCR5 is not required, as it can be substituted with the amino-terminal domains of CCR2b, murine CCR5, and CXCR2 (28, 29). This indicates that M tropic HIV-1 strains can use either the extracellular loops or the amino-terminal domain of CCR5. By contrast, dual-tropic HIV-1 env proteins are especially sensitive to amino-terminal domain substitutions and exhibit a reduced ability to use the extracellular loops of CCR5 (ref. 29; unpublished data). These observations suggest that CCR5-restricted viruses may evolve to dual tropism by acquiring the ability to recognize the extracellular loops of CXCR4, which are more closely related to their corresponding regions in CCR5, while retaining the ability to use the highly divergent CCR5 amino-terminal domain. To examine this hypothesis, we constructed two CXCR4–CCR5 chimeras, 4555 and 5444, and tested their ability to function as coreceptors for M, T, and dual-tropic env proteins. Chimera 4555 functioned as a coreceptor for the M tropic env JR-FL but not dual-tropic or T tropic env proteins (Fig. 4). By contrast, chimera 5444 functioned as a coreceptor for all M tropic, dual-tropic, and T tropic env proteins tested. These results are consistent with the hypothesis that M tropic viruses can use either the extracellular loops or amino-terminal domain of CCR5, that dual-tropic viruses are largely dependent upon the amino-terminal domain of CCR5, and that T and dual-tropic viruses interact with the extracellular loops of CXCR4 and exhibit little dependence upon the CXCR4 amino-terminal domain.
Effects of Cytoplasmic Domain Mutations on CXCR4 Coreceptor Function.
Binding of ligands to CXC chemokine receptors results in the transduction of a signaling event and mobilization of free cytosolic calcium ions, followed by receptor desensitization and internalization (40). To determine whether signaling plays a role in coreceptor function, we changed the DRY sequence in the second intracellular loop of CXCR4 to NAA (CXCR4-NAA). The DRY motif in related receptors plays an important role in G protein coupling, including CCR5 (unpublished data). To determine whether receptor desensitization and internalization is required for coreceptor function, we truncated the cytoplasmic tail of CXCR4 at residue Ser-319 and changed Ser-312 and Thr-318 to Ala residues, thereby removing the predicted phosphorylation sites in this domain. A similar truncation in CXCR2 severely impairs receptor internalization after addition of interleukin 8 (41), although substitution of alanine for serine and threonine residues in the cytoplasmic tail of CCR2b also inhibits receptor internalization (42). We found that both CXCR4-NAA and CXCR4-tail supported env-mediated cell–cell fusion (Table 1), indicating that G-protein coupling via the DRY motif and phosphorylation of the CXCR4 cytoplasmic tail are not required for coreceptor function.
DISCUSSION
The HIV-1 env protein mediates fusion between the viral and cellular membranes, allowing the viral genome to gain entry to the host cell cytoplasm (for review, see ref. 1). For membrane fusion to occur, specific conformational changes in the env protein must be triggered that are likely to result in the exposure of the hydrophobic fusion peptide at the amino terminus of the gp41 subunit. Insight into the events that trigger the fusogenic conformation, as well as the nature of this conformation, will be required to understand the mechanisms by which HIV-1 enters cells. The discovery of chemokine receptors as cofactors for HIV-1 entry provides new opportunities for studying this process. Binding to CD4 results in conformational changes in env but not in membrane fusion (1, 4). Only in the presence of an appropriate coreceptor, generally CXCR4 or CCR5, does fusion occur. For M tropic env proteins, CD4 greatly increases the efficiency of CCR5 binding (24, 25). CD4 binding may also facilitate the interaction between T tropic env proteins and CXCR4 (23). To identify regions in CXCR4 that are required for coreceptor function and that may interact with env, we examined the ability of chimeras based on CXCR4 and CXCR2, which share approximately 35% amino acid identity, to support env-mediated cell–cell fusion.
We found that the amino-terminal domain of CXCR4 did not confer coreceptor function to CXCR2 (chimera 4222) or CCR5 (chimera 4555) in contrast to what we and others have observed with CCR5 where this region imparts activity when placed in CCR2b, CCR1, CXCR2, and murine CCR5 backgrounds (26–29). Thus, although the amino-terminal domain of CCR5 is sufficient for coreceptor activity in a CXCR2 background, that from CXCR4 is not. We also found that the CXCR4 amino terminus could be substituted with the corresponding region from CXCR2 (chimera 2444b) without loss of function for four of five env proteins tested, even though these domains share little similarity at the level of primary structure. However, chimera 2444b supported fusion less efficiently than wild-type CXCR4, despite similar levels of surface expression, indicating that the amino terminus may contribute directly to the binding of env or indirectly by promoting a conformation that favors this interaction. The absolute dependence of HIV-1 RF on the CXCR4 amino terminus also indicated that this region can play a role in coreceptor function. However, the amino-terminal domain of CXCR4 appears less important than the amino-terminal domain of CCR5 for env-mediated fusion.
The inability of the CXCR4 amino-terminal domain to confer coreceptor activity to CXCR2 or CCR5 and the ability of four of the five virus strains tested to use chimera 2444b indicated that one or more extracellular loops of CXCR4 are the primary determinants of coreceptor function. Of the three extracellular loops, the third was dispensable for coreceptor function, as it could be replaced by the divergent third loop of CXCR2, which shares only 5 of 21 residues. However, as was observed with chimera 2444b, chimera 4442 supported fusion with reduced efficiency compared with wild type, indicating that the third extracellular loop of CXCR4 may directly participate in coreceptor activity to some degree, contribute to the formation and maintenance of a conformation that is permissive for the interaction with env, or modulate surface expression levels. The ability of chimera 2442 to support env-mediated membrane fusion clearly implicates the first and second extracellular loops of CXCR4 as domains critically important for coreceptor activity. Chimeras in which these domains were derived from different parental receptors revealed that the second loop was the most important determinant for coreceptor function for the HIV-1 BH8, 89.6, and BK132 env proteins. Thus, unlike CCR5, the major determinants for CXCR4 coreceptor function reside in the first and second extracellular loops.
M tropic viruses that use CCR5 as a coreceptor are responsible for viral transmission, as indicated by the remarkable resistance to infection that has been observed in individuals who lack functional CCR5 (19–22). Over the course of infection, the dynamic nature of tropism is manifested by transition of coreceptor usage by env from CCR5 to CXCR4, with dual-tropic viruses perhaps representing an important intermediate phenotype (9). Whether dual tropism is an obligate intermediate during the transition from M to T tropism is not known. The ability of dual-tropic viruses to use both CCR5 and CXCR4 offers important insights into the evolution of chemokine receptor usage in vivo. Previously, we have shown that the first 20 amino acids of CCR5 are sufficient to confer coreceptor function to diverse chemokine receptors, including CCR2b, CCR1, CXCR2, and CXCR4. However, viruses that use only CCR5 as a coreceptor can also interact with the extracellular loops and so can tolerate substitution of the CCR5 amino-terminal domain with the corresponding regions from divergent chemokine receptors (26, 29). These results suggest that CCR5-restricted viruses can interact with two sites on CCR5, one that resides predominantly in the amino-terminal domain and one that resides in the extracellular loops. In contrast to M tropic viruses, dual-tropic env proteins exhibit a reduced ability to use the extracellular loops of CCR5 and are more sensitive to mutations in the amino-terminal domain (26, 29). Our results provide an explanation for this finding: dual-tropic env proteins have acquired the ability to use the first and especially the second extracellular loops of CXCR4, while retaining the ability to use the CCR5 amino-terminal domain, even when it is placed in highly divergent backgrounds. Thus, dual-tropic env proteins such as 89.6 use different domains of CCR5 and CXCR4.
The ability of T and dual-tropic env proteins to use the first and second extracellular loops of CXCR4 rather than the amino-terminal domain may reflect the degree of similarity between the former regions and those in CCR5. The first 20 residues of CCR5 are sufficient to confer coreceptor function to divergent chemokine receptors (26, 29). The corresponding region in CXCR4 is composed of 28 amino acids, shares only three residues with CCR5, and contains an N-linked glycosylation site that is probably used (31). By contrast, the extracellular loops of CXCR4 and CCR5 exhibit a greater degree of similarity, particularly the first loop in which 7 of 14 residues are identical. Therefore, we propose that CCR5-restricted viruses become dual-tropic by acquiring the ability to use the more highly conserved, but more negatively charged, extracellular loops of CXCR4 while retaining the ability to productively interact with the first 20 residues of the much more highly divergent CCR5 amino-terminal domain. The chimeric receptors we constructed between CCR5 and CXCR4 support this hypothesis. Chimera 4555 functioned as a coreceptor for M tropic, but not dual- or T tropic, viruses. Chimera 5444 functioned as a coreceptor for all M, dual-, and T tropic viruses tested. These results are consistent with M tropic viruses using either the extracellular loops or amino-terminal domain of CCR5, dual-tropic viruses being largely dependent upon the amino-terminal domain of CCR5, and T and dual-tropic viruses interacting with the first and second loops of CXCR4.
The results presented herein, in conjunction with our earlier studies on CCR5 coreceptor activity (29), provide a framework for understanding the evolution of chemokine receptor usage by the HIV-1 env protein. Env interactions with CCR5 and CXCR4 are clearly complex and somewhat variable in nature. Although the model we have proposed to explain the evolution of chemokine receptor usage invokes env interactions with distinct CCR5 and CXCR4 domains, it is important to stress that our results are dependent upon the experimental approach and parental chemokine receptors used as donors and that chimeras between CXCR4, CCR5, and other chemokine receptors may reveal facets of coreceptor function that are not apparent from the CXCR4–CXCR2 chimeras studied herein. Furthermore, the highly conserved disulfide bonding pattern exhibited by the chemokine receptors indicates that the four extracellular domains interact closely with one another. Thus, it is not surprising that mutations in one extracellular domain sometimes influence the function of another (unpublished data). In addition, we have relied on a cell–cell fusion assay in which env, CD4, and the coreceptor are overexpressed. Although this enabled us to ascertain combinations of components that lead to membrane fusion, it will be important to revisit this issue and determine the extent to which the utilization of coreceptors is affected by the relative amounts of CD4 and chemokine receptors on target cells (26). Finally, it will be important to correlate our results with changes in env protein structure that lead to modification of the repertoire of coreceptors used, as well as in how any given coreceptor is used.
Acknowledgments
We thank Richard Duman for providing rat CXCR4, and Ben Doranz for reading the manuscript. This work was supported by National Institutes of Health Grants AI-35383, AI-38225, and AI-40880 to R.W.D. and by the Agnes Brown Duggan Endowment and the Humana Endowment to S.C.P. A number of important reagents were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. J.F.B. was supported by a Howard Hughes Medical Institute predoctoral fellowship.
References
- 1.Moore J, Jameson B, Weiss R, Sattentau Q. In: Viral Fusion Mechanisms. Bentz J, editor. Boca Raton, FL: CRC; 1993. pp. 233–289. [Google Scholar]
- 2.Wild C, Dubay J W, Greenwell T, Baird T, Oas T G, McDanal C, Hunter E, Matthews T. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sattentau Q J, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashorn P A, Berger E A, Moss B. J Virol. 1990;64:2149–2156. doi: 10.1128/jvi.64.5.2149-2156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesebro B, Buller R, Portis J, Wehrly K. J Virol. 1990;64:215–221. doi: 10.1128/jvi.64.1.215-221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. Cell. 1986;47:333–385. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 8.Clapham P R, Blanc D, Weiss R A. Virology. 1991;181:703–715. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fauci A. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 11.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 12.Bleul C C, Farazan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. Nature (London) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 15.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio P D, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 18.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 19.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 20.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 21.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kuntsman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 23.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 25.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 26.Kozak S L, Platt E J, Madani N, Ferro F E, Peden K, Kabat D. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edinger A L, Amedee A, Miller K, Doranz B J, M, Endres M S, Samson M, Lu Z-H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 29.Rucker J, Samson M, Doranz B J, Libert F, Berson J, Yi Y, Collman R G, Vassart G, Broder C C, Doms R W, Parmentier M. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 30.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, S, Choe P J V, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. Cell. 1996;87:745–765. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 31.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broder C C, Berger E A. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakrabarti S, Mizukami T, Franchini G, Moss B. Virology. 1990;178:134–142. doi: 10.1016/0042-6822(90)90386-6. [DOI] [PubMed] [Google Scholar]
- 34.Broder, C. C. & Collman, R. G. (1997) J. Leukocyte Biol., in press. [DOI] [PubMed]
- 35.Alexander W A, Moss B, Fuerst T R. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nussbaum O, Broder C C, Berger E A. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuntharapai A, Lee J, Hebert C Z, Kim K J. J Immunol. 1994;153:5682–5688. [PubMed] [Google Scholar]
- 38.Simmons G, Wilkinson D, Reeves J D, Dittman M T, Beddows S, Weber J, Carnegis G, Gesselberger U, Gray P W, Weiss R A, Clapham P R. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong M L, Xin W W, Duman R S. Mol Psychol. 1996;1:133–140. [PubMed] [Google Scholar]
- 40.Probst W C, Snyder L A, Schuster D I, Brosius J, Sealfon S C. DNA Cell Biol. 1996;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 41.Prado G N, Suzuki H, Wilkinson N, Cousins B, Navarro J. J Biol Chem. 1996;271:19186–19190. doi: 10.1074/jbc.271.32.19186. [DOI] [PubMed] [Google Scholar]
- 42.Franci C, Gosling J, Tsou C-L, Coughlin S R, Charo I F. J Immunol. 1996;157:5606–5612. [PubMed] [Google Scholar]