Abstract
During cardiac hypertrophy, the chief myocardial energy source switches from fatty acid β-oxidation (FAO) to glycolysis—a reversion to fetal metabolism. The expression of genes encoding myocardial FAO enzymes was delineated in a murine ventricular pressure overload preparation to characterize the molecular regulatory events involved in the alteration of energy substrate utilization during cardiac hypertrophy. Expression of genes involved in the thioesterification, mitochondrial import, and β-oxidation of fatty acids was coordinately down-regulated after 7 days of right ventricular (RV) pressure overload. Results of RV pressure overload studies in mice transgenic for the promoter region of the gene encoding human medium-chain acyl-CoA dehydrogenase (MCAD, which catalyzes a rate-limiting step in the FAO cycle) fused to a chloramphenicol acetyltransferase reporter confirmed that repression of MCAD gene expression in the hypertrophied ventricle occurred at the transcriptional level. Electrophoretic mobility-shift assays performed with MCAD promoter fragments and nuclear protein extracts prepared from hypertrophied and control RV identified pressure overload-induced protein/DNA interactions at a regulatory unit shown previously to confer control of MCAD gene transcription during cardiac development. Antibody “supershift” studies demonstrated that members of the Sp (Sp1, Sp3) and nuclear hormone receptor [chicken ovalbumin upstream promoter transcription factor (COUP-TF)/erbA-related protein 3] families interact with the pressure overload-responsive unit. Cardiomyocyte transfection studies confirmed that COUP-TF repressed the transcriptional activity of the MCAD promoter. The DNA binding activities and nuclear expression of Sp1/3 and COUP-TF in normal fetal mouse heart were similar to those in the hypertrophied adult heart. These results identify a transcriptional regulatory mechanism involved in the reinduction of a fetal metabolic program during pressure overload-induced cardiac hypertrophy.
Keywords: fatty acid β-oxidation, transcriptional regulation, nuclear hormone receptors, mitochondria
Fatty acid oxidation (FAO) is the major source of energy in the adult mammalian heart (1). The chief myocardial energy source switches from glucose and pyruvate in the fetal period to fatty acids after birth (1, 2). Studies in a variety of mammalian species, including humans, have shown that with cardiac hypertrophy, myocardial glucose utilization increases and FAO decreases: a reinduction of the fetal energy metabolic program (3–8). Myocardial energy utilization pathways also undergo alterations during cardiac hypertrophy, as evidenced by the re-expression of the fetal isoforms of a variety of ATPases involved in contractile function (e.g., myosin heavy chains) and ion homeostasis (e.g., Ca2+- and Na+-K+ ATPases) (9–16). Accordingly, reactivation of fetal gene regulatory programs in the hypertrophied heart may serve to reduce ATP and oxygen consumption requirements, albeit at a cost of diminished energy-producing capacity and contractile reserve. Little is known about the molecular regulatory mechanisms involved in this cardiac energy substrate switch.
Several observations suggest that reduced capacity for myocardial FAO is linked to cardiac hypertrophic growth. Humans with inborn errors in mitochondrial FAO cycle enzymes often develop ventricular hypertrophy in the absence of stimuli (such as hypertension) known to induce cardiac hypertrophy (17). Further, cardiac hypertrophy develops in rodents fed drugs that inhibit fatty acid import into the mitochondrion (18–20). Taken together with the results of studies demonstrating that fatty acid utilization is reduced during pressure overload-induced hypertrophy, these findings suggest that regulatory pathways involved in myocardial lipid metabolism are coupled to cardiac hypertrophic growth.
We recently have demonstrated that the expression of mitochondrial FAO cycle enzymes is down-regulated at the pretranslational level in failing human and rat heart (21). We sought to characterize regulation of FAO enzyme gene expression with short-term pressure overload-induced cardiac hypertrophy and to delineate the trans-acting regulatory factors involved in this response. In this report, we demonstrate that during development of pressure overload-induced ventricular hypertrophy, FAO enzyme gene expression is repressed through reactivation of fetal transcriptional control via the orphan nuclear receptor chicken ovalbumin upstream promoter transcription factor I (COUP-TF)/erbA related protein 3. Our data also implicate the transcription factors Sp1 and Sp3 in this cardiac energy metabolic gene regulatory pathway.
METHODS
Pulmonary Artery Banding (PAB) Protocol.
PAB studies were performed in adult C57BL/6 × SJL/J mice (n = 19) for 7 days as described previously (22). In brief, after induction of anesthesia and intubation, the main pulmonary artery was isolated by blunt dissection, and a suture was placed around the vessel and tied against a 25-gauge blunt needle to produce a stenosis. Sham-operated animals (n = 24) underwent the identical surgical procedure except that the constriction was not placed. After 7 days, animals from the PAB and sham groups were killed, and ventricular and atrial chambers were dissected out and snap-frozen. The mean age of the mice was 9 months with a similar distribution of male and female animals between the PAB and control groups. Animal experiments were conducted in strict accordance with the National Institutes of Health guidelines regarding humane treatment for the care and use of laboratory animals. All animal experiments were reviewed and approved by the Animal Care Committees of Washington University and the University of California at San Diego.
Northern and Protein Immunoblot Studies.
RNA isolation and Northern blot analyses were performed (21) with the cDNA probes described in Results. Total RNA was isolated from mouse left ventricle (LV) and right ventricle (RV). Protein immunoblot studies were performed as described (23), with crude nuclear protein extracts prepared from adult mouse RV or LV or pooled mouse embryonic day 16.5 hearts. The following polyclonal antibodies were used: anti-medium-chain acyl-CoA dehydrogenase (MCAD) (21), anti-long-chain acyl-CoA dehydrogenase (a gift from Arnold Strauss, Washington University), anti-Sp1 and anti-Sp3 (Santa Cruz Biotechnology), anti-COUP-TF [a gift from A. J. Butler, Imperial Cancer Research Fund, London (24)], anti-peroxisome proliferator activated receptor α [a gift from Michael Arand, University of Mainz, Germany (25)], and anti-actin (universal actin antibody, Sigma). The anti-COUP-TF antibody recognizes both COUP-TFI and COUP-TFII isoforms (24).
Transgenic Mice and Chloramphenicol Acetyltransferase (CAT) Enzyme Activity Studies.
Production and characterization of the MCADCAT.371 transgenic mice have been described (26). Pulmonary artery banding and sham procedures were performed on mice from two independent transgenic lines. CAT enzyme activities were determined on tissue extracts by use of an assay described previously (26).
Electrophoretic Mobility-Shift Assays (EMSAs).
EMSAs were performed with crude nuclear extracts prepared from pooled embryonic (day 16.5 and 19.5) mouse hearts and adult control and hypertrophied RVs, as described (26). The following sense strand nucleotide sequences represent the probes used in the EMSAs: 371/255 (the combined nuclear hormone receptor response element (NRRE)-1/site A sequence) 5′-TTGAATCCGCCAAGCAGACACGATCTGGGTTTGACCTTTCTCTCCGGGTAAAGGTGAAGGCTGACCACGGGGGCCGCTCTCCCTCCAGGCCCCAGCCACGCCCTCTAACCCAGGTTC-3′; NRRE-1, 5′-GATCCGGGTTTGA- CCTTTCTCTCCGGG; and site A, 5′-GATCCGGCCCCAGCCACGCCCTCTAACCCAG-3′ (underlined nucleotides represent partial restriction endonuclease sites added to facilitate cloning). Antibody “supershift” experiments were performed with the antibodies described above except that a different anti-COUP-TF antibody was used (a gift from Ming-Jer Tsai, Baylor University).
Cell Culture and Cotransfection Studies.
Cardiomyocytes were prepared from 1-day-old rats as described (26). Transient transfections were performed with DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) (Boehringer Mannheim) in 12-well collagen-coated dishes. For each transfection, 4 μg of reporter DNA and 2 μg of a plasmid containing a β-galactosidase gene downstream of the Rous sarcoma virus promoter were added to 15 μl of DOTAP-Hepes solution and incubated for 15 min at room temperature. In some dishes 1 μg of pCDM.COUP, a mammalian expression vector containing a human COUP-TF I cDNA downstream of a cytomegalovirus promoter (27) was added. One μg of pCDM(−), the expression plasmid backbone lacking the COUP-TF I cDNA, was added to dishes not containing pCDM.COUP. The mixture was subsequently added to the primary cardiocytes before initial plating. The cells were washed and refed the next morning. The cells were harvested 48 hr after plating. Luciferase activity was measured by the standard luciferin-ATP assay, and β-galactosidase activity was measured by the Galacto-Light chemiluminescence assay (Tropix, Bedford, MA) in an Analytical Luminescence Monolight 2010 luminometer. The reporter plasmid used for the experiments (MCADLuc.371) contains the identical MCAD promoter fragment present in the MCADCAT.371 construct fused to a luciferase reporter in the pGL2-basic plasmid (Promega).
Statistical Analysis.
Differences between values for mRNA, protein levels, CAT enzyme activities, and luciferase activities were determined with the unpaired or paired Student t tests. A statistically significant difference was defined as a P value < 0.05. All values shown represent the mean ± SEM.
RESULTS
Expression of Fatty Acid Oxidation and Glycolytic Enzymes Is Regulated at the Pretranslational Level During Pressure Overload-Induced Cardiac Hypertrophy.
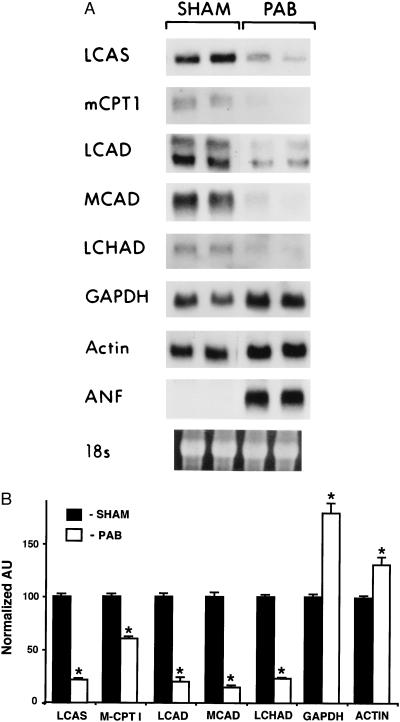
To characterize the expression of genes encoding enzymes involved in myocardial fatty acid and glucose utilization pathways, Northern blot studies were performed with RNA isolated either from the RV of mice subjected to PAB or ventricles of sham-operated controls. After 7 days of pressure overload, a significant increase (P < 0.01) was observed in mean absolute RV weight (from 29.8 ± 0.7 mg to 62.0 ± 3 mg), RV/total body weight ratio (from 96 ± 2 to 202 ± 12 × 10−5), and RV/LV weight ratio (from 0.41 ± 0.01 to 0.94 ± 0.07) in the PAB group compared with values in controls. Levels of mRNA encoding the following enzymes involved in the myocyte fatty acid utilization pathway were evaluated: long-chain acyl-CoA synthetase, the cytosolic enzyme that catalyzes the formation of fatty acyl-coenzyme A thioesters from free fatty acids entering the cell; the muscle isoform of carnitine palmitoyltransferase I, the integral outer mitochondrial membrane protein that catalyzes the initial step in mitochondrial import of long-chain acyl-CoA; and enzymes that catalyze the first (long-chain acyl-CoA dehydrogenase and MCAD) and third (long-chain 3-OH acyl-CoA dehydrogenase) steps of the mitochondrial β-oxidation cycle. For comparison, mRNA encoding the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also examined. The expression of all FAO enzyme mRNAs evaluated was significantly lower in the hypertrophied RV than in ventricles from control mice (Fig. 1A). Mean long-chain acyl-CoA synthetase and muscle isoform of carnitine palmitoyltransferase I mRNA levels were reduced to 21 ± 2% (P < 0.001) and 60 ± 2% (P = 0.004) of control levels, respectively (Fig. 1B). Mitochondrial β-oxidation cycle enzyme mRNA levels were markedly lower in the hypertrophied ventricles (long-chain acyl-CoA dehydrogenase mRNA levels were 40 ± 5% of control levels, P < 0.001; MCAD mRNA levels 15 ± 2% of control, P < 0.001; and long-chain 3-OH acyl-CoA dehydrogenase mRNA levels 23 ± 3% of control, P < 0.001). In contrast, mean GAPDH mRNA levels were 75 ± 10% higher in the pressure-overloaded RVs than in controls (P = 0.01; Fig. 1 A and B), consistent with the known increase in glucose utilization in the hypertrophied heart. As expected, expression of atrial natriuretic factor mRNA, a known marker for ventricular hypertrophy, was markedly induced in samples from hypertrophied RV. β-actin mRNA levels were modestly increased in the hypertrophied RVs compared with those in controls. Immunoblot studies demonstrated that MCAD and anti-long-chain acyl-CoA dehydrogenase protein levels paralleled the corresponding mRNA levels (data not shown). These results indicate that the expression of genes encoding cardiac fatty acid utilization enzymes is coordinately repressed at the pretranslational level, in parallel with the known reduction in myocardial fatty acid utilization during pressure overload-induced cardiac hypertrophy.
Figure 1.
Expression of genes involved in myocardial fatty acid utilization and glycolysis parallels known changes in substrate preferences in the hypertrophied heart. (A) Representative Northern blot analysis obtained with total RNA isolated from the RV of two mice subjected to PAB or from sham-operated controls (SHAM). Each lane contains 15 μg of total RNA. cDNA probe abbreviations are defined in the text. The 18S rRNA (stained with ethidium bromide) within each lane is shown at the bottom. (B) Densitometric analysis of Northern blots obtained with RNA isolated from the RV of six PAB or six sham-operated control mice. The bars represent mean (± SEM) steady-state mRNA levels (in arbitrary units or AU) normalized to sham-operated control values (mean control value set at 100). ∗ denotes P < 0.01 compared with the corresponding control value. LCAD, long-chain acyl-CoA dehydrogenase; LCAS, long-chain acyl-CoA synthetase; MCPT1, muscle isoform of carnitine palmitoyltransferase I; LCHAD, long-chain 3-OH acyl-CoA dehydrogenase.
Repression of MCAD Gene Expression During Cardiac Hypertrophy Occurs at the Transcriptional Level Through cis-Acting Sequences Within the Proximal Promoter Region.
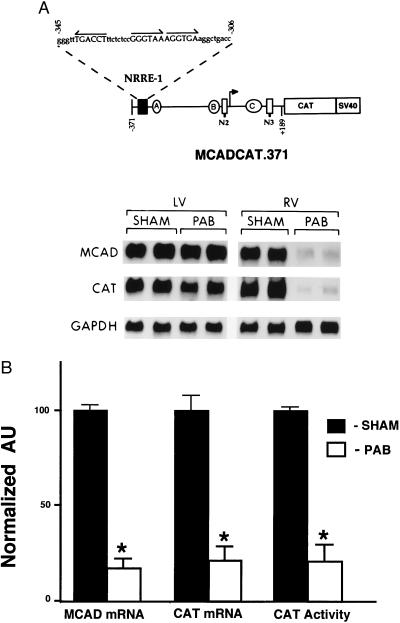
To determine whether the repression of MCAD gene expression during pressure overload-induced cardiac hypertrophy occurs at the level of transcription, PAB studies were repeated on mice transgenic for a 560-bp human MCAD promoter fragment fused to a CAT reporter (MCADCAT.371). Previous studies have shown that the expression of MCADCAT.371 in the developing murine heart parallels that of the endogenous MCAD gene (26). Specifically, expression of the MCADCAT.371 transgene is induced markedly after birth in parallel with increased postnatal myocardial fatty acid utilization rates. MCAD and CAT mRNA levels were coordinately down-regulated in the pressure-overloaded RV compared with levels in the control group and compared with those in the contralateral LV within the same animal (representative Northern blot shown in Fig. 2A). In the RV hypertrophy group, mean RV MCAD and CAT mRNA levels were 14 ± 2% (P < 0.001) and 22 ± 5% (P < 0.001) of control levels, respectively (Fig. 2B). Mean RV CAT enzyme activity in the PAB group was 22 ± 10% (P < 0.001) of corresponding control values (Fig. 2B). As expected, RV GAPDH mRNA levels were up-regulated by 73 ± 8% (P = 0.01) in the PAB group. In the LV, mean MCAD, CAT, and GAPDH mRNA levels were not significantly different in the PAB and control groups (Fig. 2A). Similar, but less pronounced, results were observed in the hypertrophied LV after aortic banding (parallel experiments, data not shown). These results indicate that repression of MCAD expression during cardiac hypertrophy occurs solely or in large part at the level of gene transcription, that the cis-acting elements involved in this transcriptional regulatory mechanism are located within the proximal MCAD promoter, and that the upstream signaling pathway involved in this energy metabolic response is triggered by the direct effect of the pressure overload stimulus.
Figure 2.
Expression of the MCADCAT.371 transgene is repressed in the hypertrophied RV. (A) The Northern blot autoradiogram is representative of the results of PAB experiments performed on mice transgenic for the MCAD promoter-CAT reporter construct MCADCAT.371 (shown schematically at the top). MCADCAT.371 includes a human MCAD gene promoter fragment (horizontal line) containing 371 bp upstream to 189 bp downstream of the transcription start site (arrow) fused to a bacterial CAT gene-simian virus 40 intron-polyadenylylation signal sequence. The positions of the three known transcriptional regulatory units in the MCAD gene promoter are represented. Each unit is composed of a NRRE (NRRE-1, N2, or N3) and a GC-rich box (A, B, or C). The sequence of potential nuclear receptor binding sites (arrows) within NRRE-1 is also shown. The Northern blot was obtained with total RNA isolated from LV and RV tissue of PAB or sham-operated control (SHAM) MCADCAT.371 mice. The blot was sequentially hybridized with cDNA probes encoding mouse MCAD, bacterial CAT, and human GAPDH. (B) Expression of the MCADCAT.371 transgene (CAT mRNA and enzymatic activity) and endogenous mouse MCAD gene (MCAD mRNA) in RV of PAB or sham-operated controls. mRNA values represent mean (± SEM) levels as determined by densitometric analysis of RNA blots obtained with total RNA from eight PAB and 10 control MCADCAT.371 mice from two independent transgenic lines [lines 10–1 (transgene copy number = 55) and 10–4 (transgene copy number = 14) described in ref. 26]. All values are shown relative to mean control values set arbitrarily at 100. Mean RV CAT enzymatic activities (normalized to mean control levels = 100) are also shown. ∗ denotes a statistically significant difference (P < 0.05) compared with controls.
The Fetal Pattern of Protein/DNA Interactions Is Reactivated at a Pressure Overload-Responsive MCAD Promoter Unit (NRRE-1/site A).
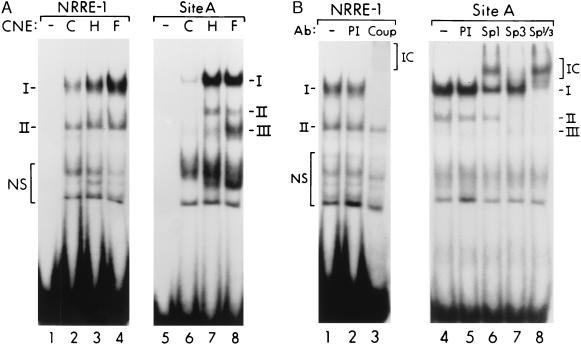
Previous studies have shown that the proximal MCAD promoter region consists of three transcriptional regulatory units, each composed of a NRRE (NRRE-1, -2, or -3) juxtaposed to a GC-rich sequence (sites A, B, or C) (elements shown schematically in Fig. 2A; ref. 28). As an initial step in the localization of the MCAD gene promoter pressure overload-responsive regions, EMSAs were performed with a series of probes derived from overlapping DNA fragments spanning the entire proximal MCAD promoter region from bp −371 to bp +189 (relative to the transcription start site = +1). EMSA performed with a probe spanning the NRRE-1/site A unit (probe 371/255, which includes human MCAD gene promoter sequence from bp −371 to bp −255, see Methods) demonstrated a significantly increased formation of DNA/protein complexes with crude nuclear protein extract prepared from hypertrophied mouse RV samples compared with sham-operated control samples (data not shown). We have shown previously that the NRRE-1/site A unit is required for appropriate cardiac developmental stage-specific expression of the MCADCAT.371 transgene (26). To further characterize the protein/DNA interactions at the NRRE-1/site A unit, separate probes containing either the NRRE-1 or site A sequence were used in EMSAs. As we have observed previously (26), the NRRE-1 probe formed two specific complexes (I and II) with cardiac nuclear proteins. Formation of complex I, but not complex II, was increased with the nuclear extracts prepared from the hypertrophied ventricle (Fig. 3A, lanes 2, 3). With the site A probe, formation of the three specific complexes (I, II, III) with the extracts derived from hypertrophied RV was markedly increased (Fig. 3A, lanes 5–7). Parallel competition studies with unlabeled specific and unrelated double-stranded oligonucleotides confirmed that complexes I and II with the NRRE-1 probe and complexes I to III with the site A probe represented specific DNA/protein interactions (data not shown).
Figure 3.
Binding of Sp proteins and COUP-TF to the MCAD promoter unit NRRE-1/Site A is induced in response to pressure overload. (A) Representative EMSAs performed with site A and NRRE-1 probes with nuclear protein extract (CNE) prepared from RV of sham-operated control mice (C), RV subjected to PAB for 7 days (H), and normal fetal (E16.5) mouse heart (F). Specific complexes are denoted by roman numerals and nonspecific interactions by NS (as defined by competition studies with unlabeled DNA fragments). (B) Antibody supershift studies performed with NRRE-1 and site A probes and nuclear protein extracts prepared from the RV of the PAB mice. Antibodies (Ab) to COUP-TF (COUP), Sp1, and Sp3 were added to the samples in lanes 3, 6, 7, and 8. Preimmune serum (PI) was added to lanes 2 and 5 as a control. Supershifted immune complexes (IC) are denoted by brackets at right margins.
The EMSAs also were performed with cardiac nuclear proteins prepared from normal embryonic day 16.5 (E16.5) mouse heart to determine whether the hypertrophy-induced protein/DNA interactions involved reactivation of the fetal DNA binding patterns. The DNA/protein interaction patterns observed with the E16.5 extracts were nearly identical to those observed with the samples from hypertrophied ventricle with either probe; bands representing complex I (with the NRRE-1 probe) and those representing complexes I, II, and III (with site A) were significantly more intense than the corresponding bands seen with the control samples from adult mice (Fig. 3A, lanes 3, 4, 7, 8). These data suggest that repression of MCAD gene expression during cardiac hypertrophic growth involves reactivation of fetal transcriptional control mechanisms at NRRE-1 and site A.
Members of the Nuclear Hormone Receptor and Sp Families of Transcription Factors Bind the MCAD Gene Hypertrophy-Responsive Unit.
To identify the proteins involved in the pressure overload-induced interactions at the NRRE-1/site A unit, antibody “supershift” studies were performed. Previous studies performed in vitro have shown that NRRE-1 is a pleiotropic NRRE that confers bidirectional transcriptional regulation via activator (e.g., retinoid X receptor α) and repressor (COUP-TF) transcription factors (27, 29). The actual nuclear receptors involved in the control of MCAD gene expression in vivo are unknown, although the results of recent antibody supershift studies with normal adult mouse heart nuclear protein extract have identified COUP-TF or a closely related protein within complex I of the NRRE-1/protein interaction (26). The putative activator(s) in complex II remain unidentified. EMSAs performed with anti-COUP-TF antibody and the NRRE-1 probe revealed that complex I, but not complex II, was supershifted by the antibody (Fig. 3B). Thus, the known transcriptional repressor COUP-TF or an antigenically related protein is indeed present within the induced complex I.
Previous in vitro studies have shown that Sp1 is capable of binding MCAD promoter site A (28). Accordingly, antibodies to multiple members of the Sp transcription factor family were used in supershift studies in an attempt to identify the proteins involved in the pressure overload-induced interactions at site A. Complex I was partially supershifted with an anti-Sp1 antibody, but not by preimmune serum (Fig. 3B, lane 6). When an anti-Sp3 antibody was used, formation of complexes I was diminished and complex II was abolished (Fig. 3B, lane 7). Addition of both anti-Sp1 and anti-Sp3 to the sample completely supershifted complex I and abolished complex II (Fig. 3B, lane 8). These results indicate that complex I contains both Sp1 and Sp3, whereas complex II contains Sp3 only. The Sp antibodies did not recognize complex III. Supershift studies with anti-Sp4 did not recognize any of the complexes (data not shown).
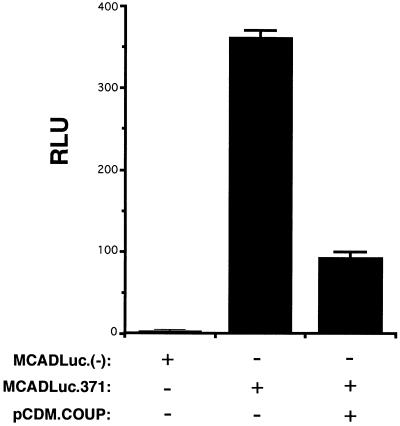
The identification of COUP-TF as one of the candidate factors responsible for the down-regulation of MCAD gene transcription in response to pressure overload is consistent with its known role as a repressor. We have shown previously that COUP-TF represses the transcriptional activity of homologous or heterologous promoters via NRRE-1 in cotransfection studies performed in several mammalian cell lines, including hepatoma G2, CV-1, and NIH 3T3 cells (27, 28). MCAD promoter elements NRRE-2 and NRRE-3 (labeled N2 and N3 in Fig. 2A) also confer modest transcriptional repression by COUP-TF (28). To confirm that COUP-TF does act to repress MCAD gene transcription in the cardiomyocyte, cotransfection experiments were performed in which primary rat neonatal cardiomyocytes were transfected with a plasmid containing the same 560-bp promoter fragment present in MCADCAT.371 fused to a luciferase reporter (MCADLuc.371) in the presence or absence of a mammalian expression vector containing a COUP-TF I cDNA (pCDM.COUP). As shown in Fig. 4, cotransfection of pCDM.COUP markedly reduced MCADLuc.371 activity (to 28 ± 4% of control). These results are consistent with those of the EMSAs in the in vivo pressure overload studies described above, and taken together implicate COUP-TF as a repressor of MCAD gene expression during pressure overload-induced cardiac hypertrophy.
Figure 4.
COUP-TF represses MCAD gene promoter activity in cardiomyocytes. The results of transient cardiomyocyte cotransfections (see Methods) are shown. The transcriptional activity of the MCAD promoter-luciferase reporter (MCADLuc.371) or luciferase reporter plasmid lacking the MCAD promoter [MCADLuc.(−)] is indicated by relative luciferase units (RLU). The bars represent mean RLU × 10−3 after correction for transfection efficiency based on the activity of cotransfected Rous sarcoma virus β-galactosidase (see Methods). Addition of the COUP-TF expression vector pCDM.COUP is indicated. The values are representative of three separate experiments.
Nuclear Levels of Sp 1/3 and COUP-TF Parallel MCAD Promoter Binding Activities in Developing and Hypertrophied Mouse Heart.
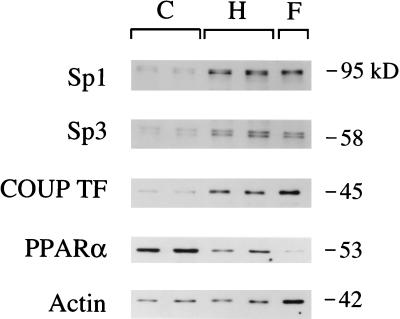
To determine whether expression of Sp1, Sp3, and COUP-TF parallel MCAD promoter binding activities during cardiac hypertrophy and in the fetal heart, steady-state nuclear levels of these transcription factors were delineated by Western blot analysis. As shown in Fig. 5, steady-state nuclear levels of Sp1, Sp3, and COUP-TF are significantly higher in hypertrophied RV and fetal heart nuclear protein extracts compared with normal adult control RV samples. In contrast, levels of anti-peroxisome proliferator activated receptor α, an orphan nuclear receptor known to induce expression of FAO enzyme genes including MCAD (29), followed a reciprocal pattern. Given that the fetal samples represented both RV and LV tissue, the immunoblot studies also were performed with nuclear proteins prepared from RV and LV of normal adult mice to address the possibility that the observed results were due to chamber-specific expression of these factors. The nuclear levels of Sp1 and COUP-TF were identical in RV and LV (data not shown), indicating that chamber-specific expression does not likely account for the differential pattern of expression observed in adult versus fetal heart samples. These data indicate that the nuclear expression of several Sp and nuclear receptor transcription factors is induced to fetal levels in response to pressure overload of the ventricle.
Figure 5.
Nuclear levels of Sp1, Sp3, and COUP-TF parallel DNA binding activities in hypertrophied, fetal, and adult heart. A representative autoradiograph of a Western blot performed with 8 μg of crude nuclear protein extract per lane. C, sham-operated control RV; H, RV hypertrophy sample; F, embryonic day 16.5 mouse heart. Protein mass markers are displayed on the right in kilodaltons (kDa). Purified Sp1 migrates on SDS-polyacrylamide gels as two polypeptide species with molecular masses of 95 and 105 kDa due to differential phosphorylation of a single polypeptide (43). The anti-Sp3 antiserum recognizes 97-, 60-, and 58-kDa polypeptides. Only the 60- and 58-kDa Sp3 signals are shown, although the expression of the 97-kDa Sp3 protein was identical to the pattern shown for the smaller isoforms (data not shown). Results with anti-PPARα and anti-actin antibodies are shown for comparison.
DISCUSSION
During the fetal stages of the developing mammalian heart, glucose and pyruvate serve as chief energy substrates (1, 2). After birth, the capacity for myocardial fatty acid oxidation increases markedly to meet the increased energy demands of the postnatal heart (30–33). During the development of cardiac hypertrophy, glycolysis increases and fatty acid utilization decreases, a reversion toward the fetal pattern of cardiac energy metabolism. Our findings demonstrate that the expression of genes involved in cellular fatty acid and glucose utilization parallel the alterations in myocardial energy substrate preferences known to occur during cardiac hypertrophy. We also demonstrate that cardiac expression of the gene encoding MCAD, which catalyzes a rate-limiting step in the β-oxidation cycle, is repressed in response to pressure overload through reactivation of fetal transcriptional control mechanisms. These results are consistent with previous reports demonstrating that pressure overload-induced cardiac hypertrophy activates expression of the fetal isoforms of genes encoding a variety of cardiac ATP-utilizing proteins, including the cardiac myosin heavy chains (15), sarcolemmal Na+, K+-ATPase (16), and the sarcoplasmic reticulum calcium ATPase (14). Thus, the paradigm of pressure overload-induced activation of the cardiac fetal gene program should be extended to include energy-producing as well as energy-utilizing pathways.
Previous studies have shown that expression of the transcription factors Fos and Jun are induced as an immediate early response to ventricular pressure overload (34–36), α1-adrenergic agonist-induced hypertrophy (37), or cardiomyocyte stretch (38). Our data identify two groups of transcription factors involved in the later stages of the hypertrophic response. To our knowledge this is the first report of a cis-acting hypertrophy-responsive element and corresponding trans-acting regulatory proteins in an in vivo preparation of ventricular pressure overload. We show that COUP-TF/erbA related protein 3 interacts with the cis-acting element NRRE-1/site A to repress MCAD gene transcription during cardiac hypertrophic growth. We also demonstrate that binding of Sp1 and Sp3 to site A is induced during cardiac hypertrophy. Although members of the Sp family may act as transcriptional activators, both Sp1 and Sp3 also have been shown to repress gene expression in a promoter context-dependent manner (39–41). Recently, we have identified a series of Sp-binding sites in the promoter region of the gene encoding the muscle isoform of carnitine palmitoyltransferase I, which catalyzes the rate-limiting step in mitochondrial fatty acid import (unpublished data), suggesting a general role for Sp proteins in the coordinate regulation of genes involved in cardiac fatty acid oxidation. Future studies will be necessary to determine whether Sp1 or Sp3 actually repress transcription of genes encoding MCAD and other FAO enzymes.
Our findings and the results of recent studies by others suggest that the transcriptional regulatory pathway described here is linked to other genes known to be regulated during cardiac hypertrophy. For example, an Sp1 binding site within the skeletal α-actin gene promoter is required for transactivation of this gene during hypertrophic growth of rat neonatal cardiocytes in cell culture studies (42). Thus, it is possible that Sp1 may function as an activator (e.g., skeletal α-actin) or repressor (e.g., MCAD) in a promoter-specific manner in response to upstream signals that trigger hypertrophic growth. Two recent reports (43, 44) indicate that nuclear receptor signaling pathways are also involved in cardiac hypertrophy programs. In these studies, the retinoid X receptor α antagonized α-adrenergic agonist- or endothelin-mediated cardiomyocyte hypertrophy in cell culture. We have shown previously that COUP-TF competes with retinoid X receptor α for binding to NRRE-1 to repress transcription (27). It is tempting to speculate that COUP-TF, as a known antagonist of retinoid X receptor, regulates the expression of many genes during cardiac hypertrophy.
Our data do not allow us to delineate the upstream regulatory mechanisms involved in the induction of COUP-TF and Sp1/3 DNA binding activities during cardiac hypertrophy. Our immunoblotting studies indicate that the nuclear levels of these factors increase during development of hypertrophy. The mechanism involved in the increased nuclear levels of these factors is unknown but could involve a post-translational modification such as phosphorylation. The results of previous studies have implicated mitogen-activated phosphorylation events in the cardiac growth response (45, 46). Recent reports have also shown that several nuclear hormone receptors, including the estrogen receptor and orphan receptor anti-peroxisome proliferator activated receptor γ are targets for mitogen-activated kinases (47, 48). Others have shown that a DNA-dependent kinase phosphorylates members of the Sp family to modify DNA binding activities (49). We speculate that growth factor signaling pathways play a role in the activation of the transcription factors described here. Future studies exploring the potential link between COUP-TF, Sp1/3, and upstream signaling pathways should prove interesting.
In summary, we have identified a gene regulatory mechanism involved in the energy substrate switch known to occur during cardiac hypertrophy. This mechanism involves reactivation of fetal transcriptional control via members of the Sp and COUP-TF/erbA related protein 3 families of transcription factors. These data and the results of previous studies suggest that this regulatory pathway is linked to other genes regulated during hypertrophic growth of the cardiac myocyte. This pathway should be a useful target for future experimental studies aimed at the characterization of the coupling of upstream signaling and transcriptional regulatory events during myocyte growth.
Acknowledgments
We thank Arnold Strauss for critical reading of the manuscript, Toni A. Rader for technical assistance with the transgenic mouse studies, and Kelly Hall for expert secretarial assistance. M.N.S. received support from a Howard Hughes Medical Institute Physician Postdoctoral Fellowship Award, and D.P.K. is an Established Investigator of the American Heart Association.
ABBREVIATIONS
- FAO
fatty acid β-oxidation
- RV
right ventricle
- LV
left ventricle
- MCAD
medium-chain acyl-CoA dehydrogenase
- COUP-TF
chicken ovalbumin upstream promoter transcription factor
- PAB
pulmonary artery banding
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility-shift assay
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- NRRE
nuclear hormone receptor response element
References
- 1.Bing R J. Harvey Lect. 1955;50:27–70. [PubMed] [Google Scholar]
- 2.Breuer E, Barta E, Zlatos L, Pappova E. Biol Neonat. 1968;12:54–64. [PubMed] [Google Scholar]
- 3.Bishop S P, Altschuld R A. Am J Physiol. 1970;218:153–159. doi: 10.1152/ajplegacy.1970.218.1.153. [DOI] [PubMed] [Google Scholar]
- 4.Taegtmeyer H, Overturf M L. Hypertension. 1988;11:416–426. doi: 10.1161/01.hyp.11.5.416. [DOI] [PubMed] [Google Scholar]
- 5.Christe M E, Rodgers R L. J Mol Cell Cardiol. 1994;26:1371–1375. doi: 10.1006/jmcc.1994.1155. [DOI] [PubMed] [Google Scholar]
- 6.Takeyama D, Kagaya Y, Yamane Y, Shiba N, Chida M, Takahashi T, Ido T, Ishide N, Takishima T. Cardiovasc Res. 1995;29:763–767. [PubMed] [Google Scholar]
- 7.Scheuer J. Circulation. 1993;87:VII-54–VII-57. [Google Scholar]
- 8.Feinendegen L E, Henrich M M, Kuikka J T, Thompson K H, Vester E G, Strauer B. J Nucl Cardiol. 1995;2:42–52. doi: 10.1016/s1071-3581(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz K, Boheler K, de la Bastie D, Lompre A M, Mercadier J J. Am J Physiol. 1992;262:R364–R369. doi: 10.1152/ajpregu.1992.262.3.R364. [DOI] [PubMed] [Google Scholar]
- 10.Alpert N R, Gordon M S. Am J Physiol. 1962;202:940–946. doi: 10.1152/ajplegacy.1962.202.5.940. [DOI] [PubMed] [Google Scholar]
- 11.Lompre A M, Schwartz K, Albis A, Lacombe B, Thiem N V, Swynghedauw B. Nature (London) 1979;282:105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- 12.Nadal-Ginard B, Mahdavi V. J Clin Invest. 1989;84:1694–1700. doi: 10.1172/JCI114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagai R, Zarain-Herzberg A, Brandl C, Fuji J, Tada M, MacLennan D H, Alpert N, Periasamy M. Proc Natl Acad Sci USA. 1989;86:2966–2970. doi: 10.1073/pnas.86.8.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Bastie D, Levitsky D, Rappaport L, Mercadier J J, Marotte F, Wisnewsky C. Circ Res. 1990;66:554–564. doi: 10.1161/01.res.66.2.554. [DOI] [PubMed] [Google Scholar]
- 15.Izumo S, Lompre A M, Matsuoka R, Koren G, Schwartz K, Nadal-Ginard B, Mahdavi V. J Clin Invest. 1987;79:970–977. doi: 10.1172/JCI112908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlemagne D, Maixent J M, Preteseille M, Lelievre L G. J Biol Chem. 1986;261:185–189. [PubMed] [Google Scholar]
- 17.Kelly D P, Strauss A W. N Engl J Med. 1993;330:913–919. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- 18.Greaves P, Martin J, Michel M C, Mompon P. Arch Toxicol. 1984;7:488–493. doi: 10.1007/978-3-642-69132-4_103. [DOI] [PubMed] [Google Scholar]
- 19.Bressler R, Gay R, Copeland J G, Bahl J J, Bedotto J, Goldman S. Life Sci. 1989;44:1897–1906. doi: 10.1016/0024-3205(89)90401-3. [DOI] [PubMed] [Google Scholar]
- 20.Rupp H, Jacob R. Eur Heart J. 1992;13:56–61. doi: 10.1093/eurheartj/13.suppl_d.56. [DOI] [PubMed] [Google Scholar]
- 21.Sack M N, Rader T A, Bastin J, McCune S A, Kelly D P. Circulation. 1996;94:2837–2842. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 22.Rockman H A, Ono S, Ross R S, Jones L R, Karimi M, Bhargava V, Ross J, Chien K R. Proc Natl Acad Sci USA. 1994;91:2694–2698. doi: 10.1073/pnas.91.7.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cresci S, Wright L D, Spratt J A, Briggs F N, Kelly D P. Am J Physiol. 1996;270:C1413–C1420. doi: 10.1152/ajpcell.1996.270.5.C1413. [DOI] [PubMed] [Google Scholar]
- 24.Butler A J, Parker M G. Nucleic Acids Res. 1995;23:4143–4150. doi: 10.1093/nar/23.20.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebel T, Arand M, Oesch F. FEBS Lett. 1992;309:37–40. doi: 10.1016/0014-5793(92)80734-x. [DOI] [PubMed] [Google Scholar]
- 26.Disch D L, Rader T A, Wood P A, Kelly D P. Mol Cell Biol. 1996;16:4043–4051. doi: 10.1128/mcb.16.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter M E, Gulick T G, Moore D D, Kelly D P. Mol Cell Biol. 1994;14:4360–4372. doi: 10.1128/mcb.14.7.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leone T C, Cresci S, Carter M E, Zhang Z, Strauss A W, Kelly D P. J Biol Chem. 1995;270:16308–16314. doi: 10.1074/jbc.270.27.16308. [DOI] [PubMed] [Google Scholar]
- 29.Gulick T, Cresci S, Caira T, Moore D D, Kelly D P. Proc Natl Acad Sci USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carroll J E, McGuire B S, Chancey V F, Harrison K B. Biol Neonat. 1989;55:185–190. doi: 10.1159/000242915. [DOI] [PubMed] [Google Scholar]
- 31.Kelly D P, Gordon J I, Alpers R, Strauss A W. J Biol Chem. 1989;264:18921–18925. [PubMed] [Google Scholar]
- 32.Nagao M, Parimoo B, Tanaka K. J Biol Chem. 1993;268:24114–24124. [PubMed] [Google Scholar]
- 33.Hainline B E, Kahlenbeck D J, Grant K, Strauss A W. Biochim Biophys Acta. 1993;1216:460–468. doi: 10.1016/0167-4781(93)90015-6. [DOI] [PubMed] [Google Scholar]
- 34.Izumo S, Nadal-Ginard B, Mahdavi V. Proc Natl Acad Sci USA. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komuro I, Kurabayashi M, Takaku F, Yazaki Y. Circ Res. 1988;62:1075–1079. doi: 10.1161/01.res.62.6.1075. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi T, Schunkert H, Isoyama S, Wei J Y, Nadal-Ginard B, Grossman W, Izumo S. J Clin Invest. 1992;89:939–946. doi: 10.1172/JCI115675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starksen N F, Simpson P C, Bishopric N, Coughlin S R, Lee W M F, Escobedo J A, Williams L T. Proc Natl Acad Sci USA. 1986;83:8348–8350. doi: 10.1073/pnas.83.21.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komuro I, Katoh Y, Kaida T, Shibazaki Y, Kurabayashi M, Hoh E, Takaku F, Yazaki Y. J Biol Chem. 1991;266:1265–1268. [PubMed] [Google Scholar]
- 39.Hagen G, Muller S, Beato M, Suske G. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okumura K, Sakaguchi G, Takagi S, Naito K, Mimori T, Igarashi H. J Biol Chem. 1996;271:12944–12950. doi: 10.1074/jbc.271.22.12944. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Hodny Z, Luciakova K, Barath P, Nelson B D. J Biol Chem. 1996;271:18925–18930. doi: 10.1074/jbc.271.31.18925. [DOI] [PubMed] [Google Scholar]
- 42.Karns L R, Kariya K, Simpson P C. J Biol Chem. 1995;270:410–417. doi: 10.1074/jbc.270.1.410. [DOI] [PubMed] [Google Scholar]
- 43.Zhou M D, Sucov H M, Evans R M, Chien K R. Proc Natl Acad Sci USA. 1995;92:7391–7395. doi: 10.1073/pnas.92.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Garami M, Cheng T, Gardner D G. J Clin Invest. 1996;97:1577–1588. doi: 10.1172/JCI118582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadoshima J, Izumo S. EMBO J. 1993;12:1681–1692. doi: 10.1002/j.1460-2075.1993.tb05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillespie-Brown J, Fuller S J, Bogoyevitch M A, Cowley S, Sugden P H. J Biol Chem. 1995;270:28092–28096. doi: 10.1074/jbc.270.47.28092. [DOI] [PubMed] [Google Scholar]
- 47.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 48.Hu E, Kim J B, Sarraf P, Spiegelman B. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- 49.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]