Abstract
DNA transposons are efficient tools in transgenesis and have therefore become popular in the analysis of the regulatory genome in vertebrates via enhancer trap screens. Here, I discuss recent progress in this field of research, with a focus on the application of one of these transposons, namely the medaka fish derived Tol2, to enhancer trapping in zebrafish, and how this approach compares with others that have a similar objective.
Introduction
TA Edison once said that, 'To invent, you need a good imagination and a pile of junk.' Noncoding DNA, representing 98.5% of the human genome, is known as 'junk' DNA and includes transposons and regulatory sequences. True to Edison's observation, the patient investigation of such junk has led to the development of transposons as vectors for DNA delivery, facilitating the study of regulatory sequences in species that are amenable to large-scale genetic analysis.
An important advantage of using transposons for insertional transgenesis is the easy identification and isolation of the DNA sequence of the genomic region surrounding the insertion site and identification of the affected gene(s). When reporter genes, promoters, or splice sites are inserted into transposons, these vectors can be used for enhancer traps (ETs) and gene traps in model animals. This approach is particularly potent when it is applied to transparent embryos, such as those of zebrafish and medaka, in which developmental events can be followed in vivo. Hence, transposons are a powerful tool in reverse genetics and developmental anatomy. Here, I focus on the use transposons for ET screens in zebrafish. Various other applications of transposons are reviewed elsewhere [1-3].
All transposons can be divided into two groups: autonomous transposons, which encode the transposase enzyme required for transposition; and non-autonomous transposons, which lack the transposase gene. Because transposase acts in trans, under normal circumstances the latter can only be mobilized in the presence of autonomous transposons. Under laboratory conditions the transposase gene can be placed onto the same DNA molecule as the transposon, supplied as another DNA molecule, or transposase activity can be provided as either mRNA or protein. Importantly, only the non-autonomous transposon, which is unable to transpose in the absence of artificially introduced transposase, can be used to develop stable transgenics. When maintenance of the transposon in the same position for many generations is a critical factor, the transposase should be introduced into the cell/embryo only briefly by injection of transposase mRNA. This is a preferred method to generate stable transgenics in species with external development of embryos, such as fish and amphibians. Until now most work has been conducted in medaka and zebrafish. Similar work using Xenopus is underway.
How to choose a transposon?
It is important to decide whether, after insertion, the transposon should remain in the same position. For gene mining, mobility of the transposon is an advantage. In contrast, in developmental studies stable transgenic lines are a must. If the model animal species used for transgenesis and the species from which the transposon originates are evolutionarily divergent, then this will assist in identifying the transposon introduced on a background of native transposons [4]. Fortunately, for workers in fish developmental biology, the bony fishes (Teleostei) represent a diverse class of vertebrates (about 25,000 species). Because molecular classification of fish is still in its infancy [5], I use zoologic classification based on morphology. Accordingly, zebrafish belongs to the Cypriniformes, which is the largest fresh water family of teleosts (about 2,000 species), and studies conducted in zebrafish could have implications for many commercial fish species, including carp, bream, and goldfish, among others.
It has been demonstrated that several transposons are capable of transposition in zebrafish [6-13]. Thus far, however, only Sleeping Beauty (SB) [9,14], Tol2 [6,10], and Ac/Ds [15,16] have been used for mass production of stable transgenics. The ET construct contains a reporter gene that encodes a fluorescent protein under the control of an attenuated promoter. When such a construct is inserted into the genome, the promoter senses the tissue-specific enhancers nearby and drives expression of the reporter gene with a particular expression pattern detected under a fluorescent dissecting microscope.
SB is the most popular transposon for animal studies. It belongs to the Tcl/mariner superfamily. It was reconstructed from a consensus sequence derived from non-autonomous Tc1-like elements of salmonid fish [17]. Salmoniformes is a family of teleosts that is evolutionarily divergent from Cypriniformes. The SB transposon contains two terminal inverted repeat/direct repeat (IR/DR) sequences required for transposition by a cut-and-paste mechanism. The synthetic SB transposon system consists of two elements: the transposase and the transposon vector containing IR/DR sequences. The recent versions of SB are much more active, suggesting that various parameters of other transposons could be improved. In medaka and zebrafish, SB has been used for ETs in zebrafish and medaka [14,18], with increased transgenesis rate compared with the plasmid injection-based approach [9].
The Tol2 transposon belongs to the hAT (hobo/Ac/Tam3) family of transposons. It has been isolated from the medaka (Oryzias latipes [Cyprinidontiformes]). The modified non-autonomous Tol2 transposes in the genome of germ cells in the presence of Tol2 transposase [8]. This methodology has been used for gene traps and ETs [6,10]. A recent application of the classical maize transposon Ac/Ds in zebrafish [15] is the subject of another review in this supplement [16].
Retrotransposons efficiently produce rearrangements of the genome, playing a major role in evolution, and their activity is linked to several human diseases [19]. Although they have attracted attention as vectors for efficient mutagenesis in the germline [20,21], they are not without drawbacks. First, a relatively rapid increase in retrotransposon copy number may compromise the maintenance of stable transgenics. Although this danger may be somewhat overestimated, a more important drawback is that retrotransposons carry cargo smaller than the size of genes encoding fluorescent proteins. Finally, retrotransposons cannot remobilize [22]. These reasons render retrotransposons inadequate for ETs.
The ET system based on a murine leukemia virus (MLV) that carries a 1 kilobase (kb) Gata2 promoter and the yellow fluorescent protein gene has been used to produce 95 zebrafish ET lines [23]. Although doubts about the bio-safety of handling such vectors could limit their use, it is also important to improve expression of fluorescent tags. It has been shown that MLV prefers to insert into the promoters of active genes [24], which in principle makes it a useful tool for generating regulatory mutants. In contrast, initial results of the ET screen have thus far failed to support this idea, showing that MLV provirus induced insertion into 5' regions of genes in only one out of eight integrations (12.5%; with a frequency five times lower than that of P-element in Drosophila [23,25]).
First lessons of enhancer trapping in zebrafish
In our laboratory, we used the non-autonomous Tol2 vector (3.2 kb) with a 1.5 kb DNA cargo, namely the EGFP gene and a basic promoter of keratin8, for a medium-sized ET screen [10]. We observed a transgenic efficiency of 16% and identified 37 transgenic lines. Thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) was used to identify DNA sequences flanking the insertion site. In most ET lines (27/37) insertions were found close to genes or within noncoding regions of genes, including introns, and 5'-untranslated and 3'-untranslated sequences (Figure 1). Eight out of 28 insertions (28.6%) were into the 5' region, which is twice as high as in the MLV screen but lower than that of P-element in Drosophila [23,25].
Figure 1.
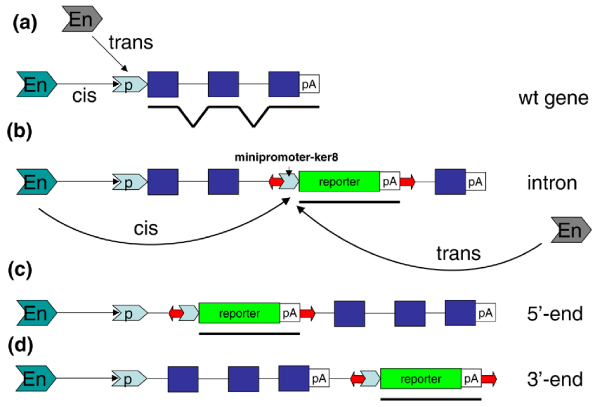
Tol2 insertion sites. Approximately half of all Tol2 insertion sites (red arrows) were (a) in close proximity to a gene, (b) in the intron, or at the (c) 5' end or (d) 3' end. The position of a hypothetical enhancer (En) is not known. The blue boxes indicate exons. p, promoter; pA, polyA.
Our construct effectively detects enhancers. In fact, characteristic tissue-specific expression of the reporter gene was found in 75% of progeny after a single cross of the founders. The positive fish was crossed again for confirmation, but we probably missed some transgenic females. At least in one case, we detected very few fluorescent embryos only every other time the F0 female was crossed. This suggested that a limited number of oogonia were transgenic because of relatively late insertion, and egg production could be a periodic process similar to that in mammals. Thus, during screening females should be crossed at least twice.
Compared with transcription, translation of GFP takes longer to reach detectable levels. Sometimes, GFP expression patterns faithfully recapitulate expression of tagged genes, for example zic3 and zic6 in ET33 [10]. In other cases, GFP expression does not correlate with expression of genes neighboring the insertion site (Garcia-Lecea and coworkers, unpublished data). This illustrates complex spatial and functional interactions between distal and proximal regulatory regions.
Although transposon activity in cis has been illustrated by analysis of expression pattern of GFP and genes tagged in ET33 (zic3 and zic6), that transposons can act in trans (Figure 1a,b) remains hypothetical. Long-range enhancer-promoter interactions have been extensively analyzed at the mammalian β-globin locus [26]. These studies suggest that communication occurs through the direct interaction of remote enhancers with the target gene by the 'looping out' of intervening chromosomal DNA (for review [27,28]), although the distal enhancers contained within the globin locus control region were not directly visualized [29]. At the same time, the long-distance interchromosomal interaction of enhancers and promoters ('transvection') has been well documented in Drosophila [30] (for review [31]). One hint that transvection may take place in vertebrates comes from experiments in which enhancers and promoters were shown to interact after co-injection on separate plasmids in a transient assay system of zebrafish embryos [32,33].
Despite some obvious limitations, the ET lines taken together reveal an endless variety of tissue-specific and cell-specific expression patterns [10]. Furthermore, the dynamic changes that occur in GFP expression pattern caused by cell migration can be followed in the same embryo in vivo for 10 to 15 hours, which in many instances provides enough time to observe the whole process of formation of individual organs (Garcia-Lecea and coworkers, unpublished data). In addition, the cytoplasmic distribution of GFP reveals even the finest cellular extensions (Figure 2).
Figure 2.
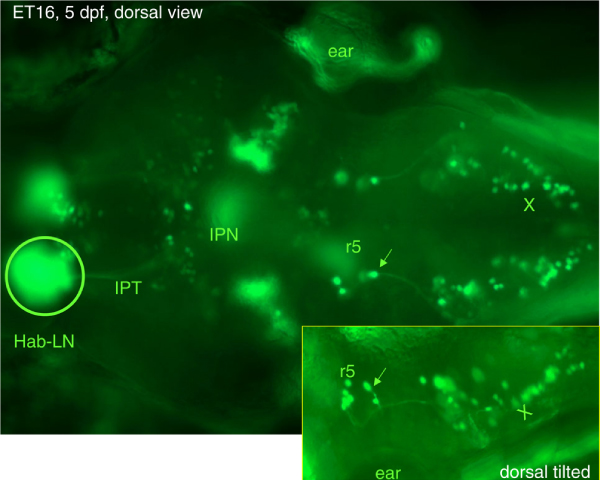
As living markers ET lines provide a possibility to analyze developmental events in vertebrates at single-cell resolution. Shown is expression of green fluorescent protein in a 5 days postfertilization ET16 larvae with an insertion of Tol2 into the 3'-untranslated region of inversin. The arrow shows a projection from r5 to the vagal nucleus. Hab-LN, lateral nucleus of habenula; IPN, interpeduncular nucleus; IPT, interpeduncular tract; r5, rhombomere 5; X, vagal nucleus.
In ET2 maternal GFP is present in all cells initially, but later on a characteristic pattern depending on the zygotic function of regulatory elements emerges. More commonly, however, expression of GFP becomes robust by the end of the first day of development, making these lines good tools for the study of organogenesis. Unfortunately, background expression in the skin is sometimes high. To alleviate this problem, embryonic skin can be removed or embryos can be sectioned optically using confocal microscopy.
In general, it is relatively simple to generate several dozen ET transgenics and characterize insertion sites, which can be accomplished by trainees. Although expression patterns in some lines are relatively easy to understand, analysis of lines with complex patterns of expression requires experienced personnel. Thus, it could be advantageous to initially develop ET lines with robust expression of a marker gene and a favorable ratio of signal to noise, and only then become involved in studying the details of expression patterns.
Study of zebrafish anatomy has been inadequate. ET lines could be used for identification and analysis of organs that have not yet been described. For example, the miniscule corpuscles of Stannius (CT), consisting of only a few cells, have been detected in association with the posterior pronephric ducts in ET2 and ET7 larvae [10]. In teleosts, CT performs some functions of the parathyroid gland. Later on other molecular markers have been linked to these structures [34]. Thus, ET lines are excellent tools for conducting detailed anatomic studies in vivo.
This analysis could be extended by crossing different ET animals expressing the same marker or ET animals expressing different markers. Such approaches could be informative in coordinated analyses of different cell lineages, organelles, or cell compartments. For now, in our laboratory we have demonstrated proof-of-principle of this approach for different cell lineages in neuromasts of the lateral line after crossing ET4 and ET20 lines [10]. This approach would greatly benefit from availability of transgenics that express other fluorescent proteins [23].
All information about ET lines generated in this laboratory has been consolidated in the database of Zebrafish Enhancer-TRAP (ZETRAP) lines [35,36], which contains a brief description of each line (expression patterns at 4 to 5 days postfertilization and sequence flanking insertion site). An evaluation of the frequency of requests revealed that the ET lines with relatively simple expression patterns were requested more often. These are, for example, ET16 that reveals an asymmetry of the habenular nuclei (Figure 2) or ET4 with expression of GFP in mechanoreceptors of the neuromast and ET20 with expression in another cell lineage of the neuromast - support glial cells. However, as we progress in our understanding of more complex patterns of expression, we expect that these more complex lines will gain popularity as well. At the time of writing, ET lines described in the ZETRAP database or plasmids have been distributed to more than 30 laboratories in 12 different countries. This will result in the generation of many novel ET lines and other transposon-based applications.
The number of transgenic lines is increasing rapidly. Some ET lines are used as 'launching pads' for transposon jumps into new sites by injection of transposase mRNA into embryos [10]. When initiating such a project, it is important to ensure that, after injection of transposase mRNA, the ectopic expression of a reporter appears in somatic cells. In the absence of such events, it is not advisable to continue because the efficiency of germline transposition will probably be much too low.
Although some expression patterns in new lines ('transposants') will be novel and unrelated to the maternal expression pattern, some transposants could represent variations of the original pattern. Also, if the expression pattern in the maternal line is complex, then the transposants with similar but simpler expression patterns could be useful in deciphering the complex pattern in the maternal line. At the DNA level, insertion of Tol2 is usually accompanied by the 8 base pair target site duplication, which often remains after excision of the transposon. The footprint sequence left after transposon relocation varies, indicating that DNA repair probably involves non-homologous end-joining, which in turn opens the possibility of mutation at the site of excision.
Recently, the size of the Tol2 transposon was substantially reduced to form the mini-Tol2, which consists of only about 0.35 to 0.50 kb of flanking sequences of the original Tol2 transposon DNA. Nevertheless, it can carry at least 10 kb of cargo without a decrease in the rate of transgenesis [37,38]. Importantly, mutation analysis of the subterminal regions of terminal inverted repeats revealed short repeated sequences essential for transposition [38]. Being able to deliver large DNA inserts, these 'mini' Tol2 vectors are of obvious interest to the gene therapy community. However, it remains unclear whether they could be used for ET screens.
Comparative analysis of promoters used in enhancer trap screens
Several different promoters have been used in ET screens in medaka and zebrafish, including regulatory regions associated with genes that are expressed ubiquitously (cska [cytoskeletal actin] of Xenopus borealis and ef1α [elongation factor 1α] [14,18], developmental regulatory genes (gata2) [23], and cell lineage-specific genes (keratin8) [10]. Application of all of these constructs resulted in the generation of transgenic lines characterized by a diverse range of expression patterns that vary from relatively ubiquitous expression to those that are tissue specific. Thus, all promoters used exhibited no preference for derivatives of any specific germ layer. It has been proposed that the generation of transgenics with tissue-specific expression patterns using the ubiquitously active promoters of cska and ef1α may reveal the activity of negative regulatory elements (silencers) [39]. For now at least, this hypothesis remains untested.
These and other details of comparison of several ET screens in medaka and zebrafish are summarized in Table 1. To ascertain the full potential of different transposon vectors, more analyses using different constructs will be necessary.
Table 1.
Comparative analysis of enhancer trap screens
| Model animal | Vector | Reporter | Promoter | Insertion bias | Insertion ite | Remobilization of vector | Footprint | Mutation | Tissue-specific pattern | Number of lines | Ref. |
| Medaka fish (Oryzeas latipes) | SB10 | GFP | cska (ubiquitous) | NA | TA | Possible | NA | NA | + | 174 | [17] |
| Zebrafish (Danio rerio) | Tol2 | GFP | Mini-keratin8 (tissue specific) | ≥ 50% close to/inside genes | Variable | Possible, shown | Variable | Possible | + | 37 | [10,35] |
| Zebrafish | SB10 | GFP | ef1α (ubiquitous) | ≥ 50% close to/inside genes | TA | Possible, shown | NA | Possible | + | 9 | [13] |
| Zebrafish | MLV provirus | YFP | gata2 (tissue specific) | Close to/inside genes | Variable | Not possible | NA | About 5% to 10% of insertions | + | 95 | [22] |
GFP, green fluorescent protein; MLV, murine leukemia virus; NA, not assessed; SB, Sleeping Beauty; YFP, yellow fluorescent protein.
Finding regulatory regions
Given the ease of identifying insertion sites, one could use transposons to map regulatory regions in the vertebrate genome and eventually identify specific regulatory elements using several complementary approaches. A computer-based approach searches for regions of noncoding DNA that are conserved between fish and humans [40]. However, because of the relatively low level of conservation of these regions at long evolutionary distances, the usefulness of this approach may be limited. Thus, given the ease of experiments that involve transient expression in zebrafish, a functional approach has been developed. Here, the regions of any DNA that may contain specific regulatory sequences, in combination with a marker gene under the control of a basic promoter, could be rapidly evaluated for tissue-specific expression after injection into zebrafish embryos [32,33,41-43]. One further approach includes the comparison of sequences of candidate regulatory regions between zebrafish and two pufferfish species whose genome sequences are already available, namely fugu (Takifugu rubripes [44]) and the spotted green pufferfish (Tetraodon nigroviridis [45]). Systematic application of all these approaches will be crucial for rapid identification of regulatory sequences and will help to put a significant pile of 'junk' DNA to better use. Once this is accomplished, it will remain to be seen whether we possess enough imagination to become inventors of applications based on emerging knowledge about the regulatory genome.
Conclusion
This review highlighted some problems of application of transposon technology that emerged as a result of several completed and ongoing enhancer trap screens based on this technology using zebrafish, which were completed in the author's laboratory. These include a selection of a suitable transposon and a regulatory region to drive expression of a marker gene, an identification of genes regulated by detected enhancer and, finally, an identification of the enhancer. Because we are at the very beginning of application of this technology in developmental biology of vertebrates, one could expect to see fast progress in this field resulting in emergence of new transgenic lines to be used as living markers of diverse cell lineages and organs. This in turn will change modern developmental biology, transforming it into a science whose findings will be validated by results of in vivo investigation.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgements
The author is thankful to two anonymous reviewers. Dr Sergey Parinov provided photographs. Drs Mark Featherstone and Lee-Thean Chu carefully read and commented on the manuscript. The author's laboratory at the Institute of Molecular and Cell Biology is supported by a grant from the Agency for Science, Technology and Research (A-STAR) of Singapore.
This article has been published as part of Genome Biology Volume 8, Supplement 1, 2007: Transposons in vertebrate functional genomics. The full contents of the supplement are available online at http://genomebiology.com/supplements/8/S1.
References
- Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- Miskey C, Izsvak Z, Kawakami K, Ivics I. DNA transposons in vertebrate functional genomics. Cell Mol Life Sci. 2005;62:629–641. doi: 10.1007/s00018-004-4232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman S, Clark K, Hackett PB. Fishing for answers with transposons. Mar Biotechnol (NY) 2005;7:135–141. doi: 10.1007/s10126-004-0068-2. [DOI] [PubMed] [Google Scholar]
- Weiser KC, Justice MJ. Sleeping Beauty awakens. Nature. 2005;436:184–186. doi: 10.1038/436184a. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Orti G, Meyer A. Novel evolutionary relationship among four fish model systems. Trends Genet. 2004;26:424–431. doi: 10.1016/j.tig.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Raz E, van Luenen HG, Schaerringer B, Plasterk RH, Driever W. Transposition of the nematode Caenorhabditis elegans Tc3 element in the zebrafish Danio rerio. Curr Biol. 1998;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AE, Balciunas D, Mohn D, Shaffer J, Hermanson S, Sivasubbu S, Cliff MP, Hackett PB, Ekker SC. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Hartl DL, Dowling JE. Transposition of the mariner element from Drosophila mauritiana in zebrafish. Proc Natl Acad Sci USA. 1998;95:5182–5186. doi: 10.1073/pnas.95.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo M, Muller F, Kiss J, Balduf C, Strahle U, Olasz F. Transposition and targeting of the prokaryotic mobile element IS30 in zebrafish. FEBS Lett. 2003;550:46–50. doi: 10.1016/s0014-5793(03)00814-7. [DOI] [PubMed] [Google Scholar]
- Miskey C, Izsvak Z, Plasterk RH, Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Gao Y, Naqvi NI, Parinov S. Trans-kingdom transposition of the maize Dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Emelyanov A. Transposable elements in fish functional genomics: technical challenges and perspectives. Genome Biol. 2007;8(Suppl 1):S6. doi: 10.1186/gb-2007-8-s1-s6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Grabher C, Henrich T, Sasado T, Arenz A, Wittbrodt J, Furutani-Seiki M. Transposon-mediated enhancer trapping in medaka. Gene. 2003;322:57–66. doi: 10.1016/j.gene.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–60. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bestor TH. Transposons reanimated in mice. Cell. 2005;122:322–325. doi: 10.1016/j.cell.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Ellingsen S, Laplante MA, Konig M, Kikuta H, Furmanek T, Hoivik EA, Becker TS. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Bellen HJ. Ten years of enhancer detection: lessons from the fly. Plant Cell. 1999;11:2271–2281. doi: 10.1105/tpc.11.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- Grosveld F, de Boer E, Dillon N, Gribnau J, McMorrow T, Milot E, Trimborn T, Wijgerde M, Fraser P. The dynamics of globin gene expression and position effects. Novartis Found Symp. 1998;214:67–79. doi: 10.1002/9780470515501.ch5. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Lopping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai Y-F, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Ronshaugen M, Levine M. Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev Cell. 2004;7:925–932. doi: 10.1016/j.devcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- Müller F, Williams DW, Kobolak J, Gauvry L, Goldspink G, Orban L, Maclean N. Activator effect of coinjected enhancers on the muscle-specific expression of promoters in zebrafish embryos. Mol Reprod Dev. 1997;47:404–412. doi: 10.1002/(SICI)1098-2795(199708)47:4<404::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Müller F, Chang B, Albert S, Fischer N, Tora L, Strähle U. Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development. 1999;126:2103–2116. doi: 10.1242/dev.126.10.2103. [DOI] [PubMed] [Google Scholar]
- Chong SW, Nguyen TT, Chu LT, Jiang YJ, Korzh V. Zebrafish id2 developmental expression pattern contains evolutionary conserved and species-specific characteristics. Dev Dynam. 2005;234:1055–1063. doi: 10.1002/dvdy.20625. [DOI] [PubMed] [Google Scholar]
- Choo BG, Kondrichin I, Parinov S, Emelyanov A, Go W, Toh WC, Korzh V. Zebrafish transgenic Enhancer TRAP line database (ZETRAP). BMC Dev Biol. 2006;6:5. doi: 10.1186/1471-213X-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebrafish Enhancer TRAP lines database http://plover.imcb.a-star.edu.sg/~zetrap/ZETRAP.htm
- Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts AM, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Lenhard B, Becker TS. New technologies, new findings, and new concepts in the study of vertebrate cis-regulatory sequences. Dev Dynam. 2006;235:870–885. doi: 10.1002/dvdy.20659. [DOI] [PubMed] [Google Scholar]
- Lee AP, Koh EGL, Tay A, Brenner S, Venkatesh B. Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proc Natl Acad Sci USA. 2006;103:6994–6999. doi: 10.1073/pnas.0601492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Blader P, Strähle U. Search for enhancers: teleost models in comparative genomic and transgenic analysis of cis regulatory elements. Bioessays. 2002;24:564–572. doi: 10.1002/bies.10096. [DOI] [PubMed] [Google Scholar]
- Plessy C, Dickmeis T, Chalmel F, Strähle U. Enhancer sequence conservation between vertebrates is favoured in developmental regulator genes. Trends Genet. 2005;21:207–210. doi: 10.1016/j.tig.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fugu rubripes v4.0 http://genome.jgi-psf.org/Takru4/Takru4.home.html
- Tetraodon Genome Browser http://www.genoscope.cns.fr/externe/tetranew/


