Abstract
Following the descovery of its transposition activity in mammalian culture systems, the Sleeping Beauty (SB) transposon has since been applied to achieve germline mutagenesis in mice. Initially, the transposition efficiency was found to be low in cultured systems, but its activity in germ cells was unexpectedly high. This difference in transposition efficiency was found to be largely dependent on chromosomal status of the host genomic DNA and transposon vector design. The SB transposon system has been found to be suitable for comprehensive mutagenesis in mice. Therefore, it is an effective tool as a forward genetics screen for tagged insertional mutagenesis in mice.
Introduction
With the completion of the mouse genomic sequencing project, efficient functional genomics is the current and most pressing focus for achieving a better understanding of gene function. Emerging transposon systems have successfully been applied to both germline and somatic cell mutagenesis in mice. These transposon systems have proven useful and should be considered essential tools for functional genomics in mice and other species
Resurrecting the DNA-type Sleeping Beauty transposon and demonstrating its transpositional activity in mammalian cells
In 1997, Ivics and coworkers [1] reported the important finding that Tc1/mariner type DNA transposase reconstructed from the salmon fish genome had significant transposable activity in mammalian cultured cells. They fittingly named the transposon 'Sleeping Beauty' (SB), because an inactive transposase has been awakened from millions of years of evolutionary sleep. Tc1/mariner superfamily transposases are known to be active in a wide range of species, ranging from protozoa to mammals. However, other members, such as insect (Himar1 and Mos1) and worm (Tc1 and Tc3) transposons, were shown to be much less active in mammalian cultured cells [2]. Initially, the obvious applications of the transposon included its use in germline mutagenesis. However, in the subsequent year Luo and colleagues [3] reported that chromosomal SB transposition was not efficient in embryonic stem cells, describing an approximate transpositional frequency of about one in 104 cells. If this efficiency is similar to that of germline transposition, then one can easily predict that the efficiency in generating mutant mice would be approximately one mutant out of 104 newborn mice, which is not suitable for applications in high-throughput forward genetics.
Strategy for detecting germline transposition in mice
Although the efficiency of chromosomal transposition was found to be low in culture systems, we wished to determine the actual germline transposition efficiency in vivo. Therefore, using a noninvasive and sensitive technique, the green fluorescent protein (GFP) reporter system was selected to trace the expression of a transgene (transposition events) [4]. As shown in Figure 1, two transgenic lines bearing the SB transposon vector containing the GFP expression cassette and expressing the SB transposase were generated. When generating transgenic mice bearing the SB transposon vector, the flanking components of the transposon vector, such as resistance genes (ampicilin and neomycin cassettes) and plasmid backbone, were also included to suppress GFP expression in the original donor concatemer. Transposition from the original donor sites would, in turn, activate GFP expression when the transposon was reintegrated into a different locus. Therefore, GFP fluorescence would indicate that SB mediated transposition events had taken place. Using this strategy, the efficiency of in vivo transposition can be examined at high sensitivity.
Figure 1.
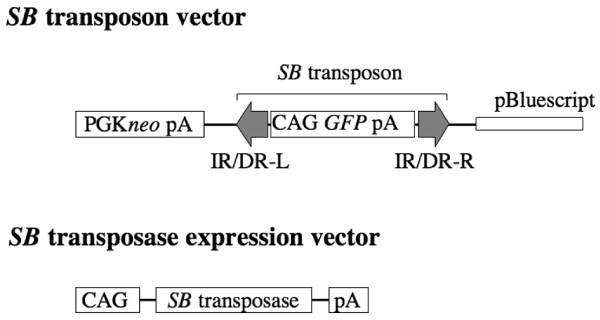
Constructs for generating the two transgenic mice. Sleeping Beauty (SB) transposon vector contains a green fluorescent protein (GFP) expression cassette flanked by SB transposon elements: inverted repeat/direct repeat (IR/DR)-L and IR/DR-R. SB transposase is driven by the CAG promoter. pA, poly(A) addition signal.
Two transgenic lines were independently produced with injections of respective construct into fertilized eggs and inter-crossed to generate double transgenic mice (Figure 2a) [4]. Approximately 20 copies of transgene were identified in the mouse bearing SB transposon at chromosome locus 3H1-H2. SB transposase gene, driven by a ubiquitous strong promoter, was inserted at chromosome locus 4C4-C5 [4]. GFP expression was carefully evaluated in somatic cells derived from the double transgenic mice by fluorescence activated cell sorting, but no GFP positive cell was identified (Figure 2b). However, evidence of SB transposon excision was observed at very high frequencies in tail DNA (approximately one excision in 1.5 cells) from the double transgenic mice [4]. SB transposon also leaves a unique 'footprint' at the excision sites, which consists of a pair of TA dinucleotides spaced by three nucleotides (CA/TG).
Figure 2.
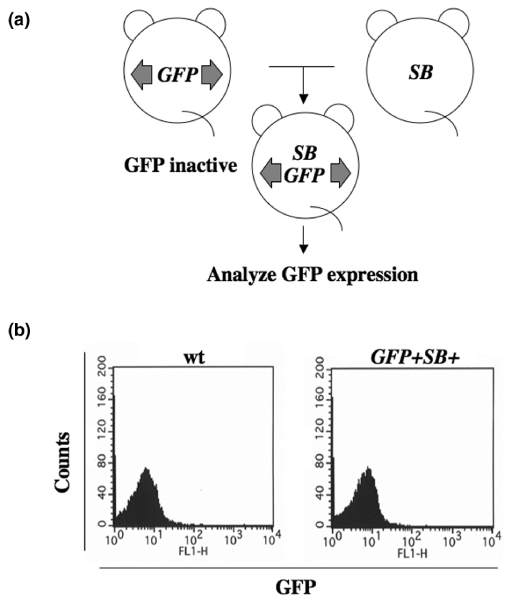
Generation and expression of GFP in double transgenic mice. (a) Transgenic mice bearing Sleeping Beauty (SB) transposon vector (green fluorescent protein [GFP] mice, left) were mated with transgenic mice expressing SB transposase (SB mice, right) to generate double transgenic mice. (b) No GFP signal was detected in peripheral blood by fluorescence activated cell sorting analysis. wt, wild-type.
The apparent discrepancy between no detectable GFP expression and high frequency of transposon excision in double transgenic mice may be reconciled by the following two possibilities (Figure 3). First, although the excision (the first step of transposition) was high in mice, the integration (the second step of the transposition) may have been extremely low (Figure 3; scheme I). This is reminiscent of RAG recombinase, which catalyzes V(D)J recombination in T and B lymphocytes. Significant similarities between RAG recombinase and Hermes transposase, classified as part of the hAT superfamily, have been reported based on the observation of hairpin intermediate formation at the ends of the donor double strand breaks [5]. RAG recombinase has transposase activity in vitro [6,7], but excised fragments during recombination are rarely integrated back into the mammalian genome in vivo. Similarly, the reintegration mechanism mediated by SB transposase could in some way be inhibited in vivo. Second, although the transposition frequency is high, detection of GFP may be problematic for unknown reason(s) (Figure 3; scheme II).
Figure 3.
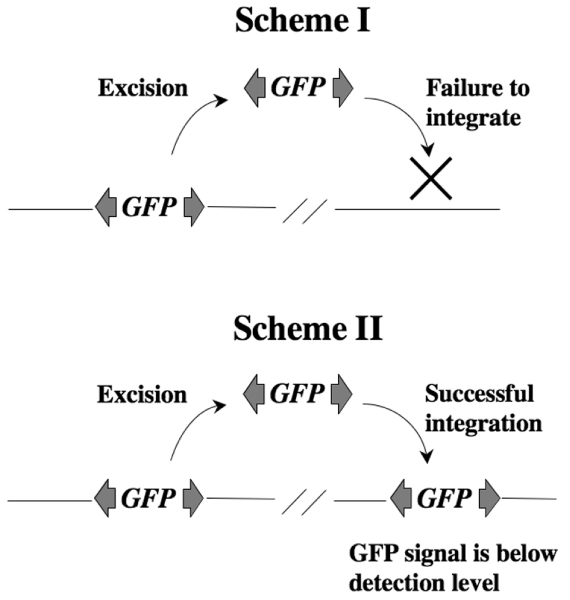
Two possible schemes to account for the absence of GFP signal in double transgenic mice. In scheme I the integration step is extremely inefficient compared with the excision step. In scheme II both integration and excision steps are efficient, but the green fluorescent protein (GFP) signal is below the detection level, or suppression of the GFP gene is maintained at the new integration site.
If transposition events occur in germ cells as well as somatic cells, then we would be able to analyze transposition sites of mice in the next generation using methods independent of GFP expression, such as Southern blot analysis, fluorescence in situ hybridization, or direct determination of integration sites using the transposon sequences as a tag. To tackle this issue, a new mating strategy was established (Figure 4). Unexpectedly, many GFP positive mice were obtained using this new mating scheme (Figure 4) [4]. Moreover, the intensity of GFP expression varied tremendously, suggesting that transposition events occur randomly and frequently in the germ cells of double transgenic mice. Fluorescence in situ hybridization analyses, using mitogen stimulated spleen cells derived from GFP positive mice, revealed the presence of novel signals that were different from the signal at the donor site (chromosome locus 3H1-H2), indicating that transpositions occurred in these mice. Germline SB transposition was also reported by two other groups using a similar mating scheme [2,8], demonstrating the validity of our observations and conclusions.
Figure 4.
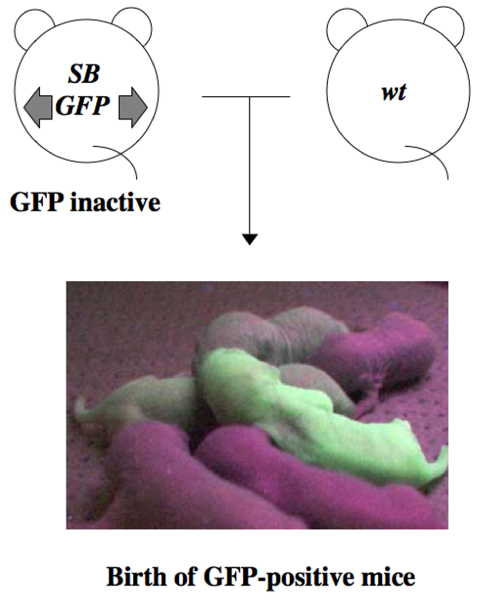
GFP expression in progeny of double transgenic mice. Double transgenic mice were mated with wild-type mice to test transmission of Sleeping Beauty (SB) transposon insertions to progeny. If transmission of SB transposon insertions occur in progeny, then it would enable us to detect insertion with methods other than green fluorescent protein (GFP) expression, such as Southern blot analysis or fluorescence in situ hybridization analysis. Some of the progeny from the double transgenic mice are clearly GFP positive, and the intensity of GFP varied, suggesting that SB transposase mediated transpositions occurred and transposons integrated at various locations. wt, wild-type.
Why could we not detect any GFP expression in the double transgenic mice? The following scheme is postulated to explain this issue (Figure 5). In the double transgenic mice transposon DNA at the original donor sites was highly methylated, and this may explain the suppression of GFP expression at the donor site. DNA methylation is mediated by DNA methyltransferases. Among these, DnmtI methylates CG dinucleotides in a template dependent manner. We speculate that the transposed DNA is also methylated by DnmtI and is continuously suppressed (Figure 5). This suppression may be abrogated during germline transmission, and the same methylation pattern would be reintroduced at the donor site in progeny. However, methylation may not be introduced to the transposed DNA, resulting in GFP expression (Figure 5).
Figure 5.
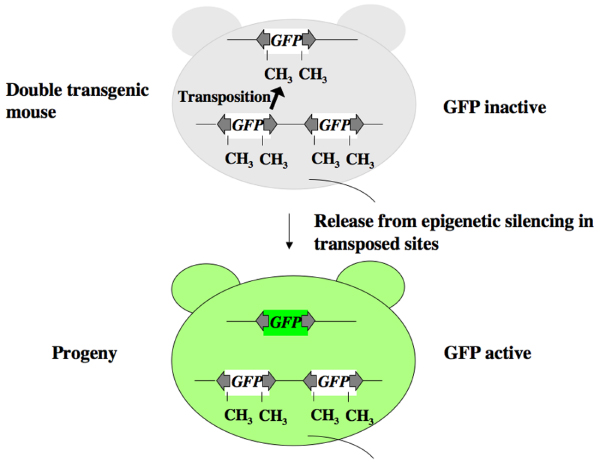
Possible explanation for why we could not detect GFP signal in double transgenic mice. Green fluorescent protein (GFP) expression at the donor site may be suppressed because of highly methylated transposon DNA (top panel). This suppression may be abrogated during germline transmission. However, the same suppression would be reintroduced at the donor site, probably because of the flanking sequence of the transposon DNA. The suppression may not be reintroduced into the transposed DNA (bottom panel).
Efficient transposition in germ cells
According to three published reports on germline SB transposition [2,4,8], transpositional frequency of germ cells was approximately 0.1 transposition/transposon per germ cell, which was about 1,000-fold higher than that of cultured cells. If the original donor site contains ten transposon copies, then each germ cell bears approximately one transposition site. Therefore, the frequency of transposition in germ cells is suitable for comprehensive mutagenesis in mice.
The other important issue would be how many different mutant mice can be generated from a double transgenic mouse. To determine the complexity, we directly analyzed spermatozoon DNA from the double transgenic mice. Because individual transposon integration sites were detected at the frequency of one in 10,000 spermatozoon and each spermatozoon contained approximately one transposed DNA, the complexity of transposition in germ cells was approximately 10,000 [9]. This means that 10,000 different mutant mice could be produced from a single double transgenic mouse, which represents a sufficiently large number of transposition events and renders this technology suitable for use in large scale forward genetics research.
Heterochromatin status affects transposition efficiency
That higher transposition frequencies were observed in mice compared with culture systems may be explained by the epigenetic status of transposons. To investigate this issue further, methylated or unmethylated transposon DNA was introduced into cultured cells [10]. The methylation status of both transposon vectors used was confirmed after integration into the genome; SB transposase was subsequently introduced and rates of excision frequency were determined [10]. Excision frequency was enhanced approximately 100-fold using methylated transposon DNA compared with unmethylated DNA. Methylated transposon DNA also introduced histone modifications such as histone H3-K9 tri-methylation, which is a marker of heterochromatin status [10].
Recently, the influence of heterochromatin conformation on SB transposition was further examined by recruiting a tetracycline-controlled transrepressor (tTR), which induces heterochromatin conformation, to the SB transposon through a tet operator sequence [11]. Chromatin immuno-precipitation analysis revealed that SB transposase was clearly bound to the tTR induced heterochromatin conformation. The frequency of SB transposition was simultaneously enhanced by approximately 100-fold. These findings indicate that the high affinity of SB transposase for heterochromatin conformation results in enhanced transposition efficiency.
Construction of Sleeping Beauty transposon vector for germline mutagenesis
We found that more than half of transposon integration sites were mapped to the same chromosome bearing the original donor site (transposon concatemer), and preferential transposition occurring near to the original donor site has been clearly demonstrated [9]. Remaining transposition events were widely and randomly distributed throughout the mouse genome, with no apparent preferential integration sites [9]. Therefore, SB transposon system can be utilized for both region specific and genome wide germline mutagenesis.
The high degree of complexity and efficiency of transpositions in our seed mice led us to apply the SB transposon system to large-scale mutagenesis in mice. For this, we designed a new version of the transposon vector bearing both promoter and poly(A) trap cassettes [9]. The original version was designed to detect transpositions irrespective of integrations into endogenous genes. The new version uses poly(A) trap and promoter trap strategies to select transposition events occurring in endogenous genes and to identify the expression pattern of trapped endogenous genes (Figure 6). Comparison of GFP intensities between the original version and poly(A) trap cassette revealed that the former mostly resulted in much greater intensity. This may partly be caused by the degradation of GFP mRNA used for poly(A) trapping via an mRNA surveillance mechanism called nonsense-mediated mRNA decay. Shigeoka and coworkers [12] reported that a specific secondary structure, such as the internal ribosome entry site (IRES) inserted 5' of the splice donor, prevented nonsense-mediated mRNA decay. Addition of IRES just upstream of the splice donor may further improve the ability of the transposon vector to trap endogenous genes. Recently, many other fluorescent proteins (cyan fluorescent protein, yellow fluorescent protein, and monomeric red fluorescent protein) have been successfully applied to allow visualization in vivo [13-15]. It has been suggested that multicolor imaging in vivo will facilitate better understanding of biologic phenomena [16]. Application of such reporter genes in new transposon vector systems may allow for more efficient tracking of trapped endogenous genes in a noninvasive and real-time manner.
Figure 6.
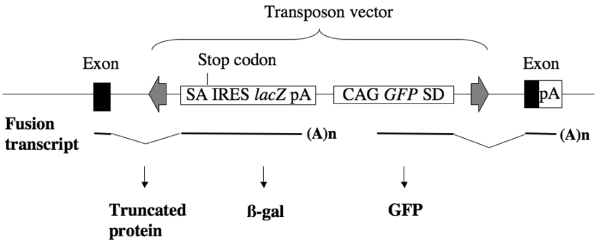
A new version of transposon vector for gene trapping. Transcription of an endogenous gene is disrupted and β-galactosidase (β-gal) is expressed. Transcription by the CAG promoter generates a chimeric transcript of the green fluorescent protein (GFP) and the endogenous gene. IRES, internal ribosome entry site; pA, poly(A) addition signal; SA, splice acceptor; SD, splice donor.
Generation of mutant mice using the Sleeping Beauty transposon system
Using a new version of the transposon vector, GFP is activated only if the transposed transposon vector is reintegrated into endogenous genes in a sense orientation. Two different transgenic mice bearing this transposon vector were generated; one has a 20-copy transposon concatemer on chromosome 12 and the other has approximately 100 copies on chromosome 7. Approximately 7% of newborn mice from both double transgenic mice derived from these two different transposon bearing mice were GFP positive, suggesting that copy number and chromosomal location of the transposon are not the major determinants of transposition efficiency. This noninvasive GFP examination allowed us to focus on potential mutants soon after birth. Mutant mice obtained from two different donor sites were extensively analyzed. It was found that region specific saturation mutagenesis was possible within a 4 megabase region of the original donor site, and remaining transpositions were widely and randomly distributed, as indicated by the first version of the transposon vector [17]. We compiled a database of SB transposon insertion sites identified in GFP positive progeny and germ cells from double positive transgenic mice [17]. This database is available online [18] and sperm from many of these lines were stocked in liquid nitrogen.
Accordingly, there are two different strategies that may be used to generate mutant mice with the SB transposon (Figure 7). First, because each donor site allows disruption of endogenous genes to occur with a region specific, saturated manner within 4 megabases, production of many donor sites distributed throughout the mouse genome may result in comprehensive generation of mutant mice in a genome-wide saturated manner (Figure 7, left). However, the drawback of region specific mutagenesis is the original donor site effect. Because the majority of transposition events occur locally, segregation between new insertion sites with local hopping and the original donor site is not easy. Also, in the case of phenotype analyses, we cannot ignore the effect of the original donor site, which may contribute to the phenotype. To address this issue, we usually screen for the donor site effect to determine whether mice homozygous for the donor sites have any apparent phenotype, and we only use transgenic mice bearing SB transposon with no phenotype [17]. In fact, we found that approximately half of mice homozygous for the donor sites were phenotype positive; these animals were not used for further experimentation. More recently, Geurts and coworkers [19] reported that there are many cases of genomic rearrangements, including translocations and deletions, occurring near the original donor sites during SB transposition events. This indicates that one must be careful when evaluating phenotype and consider whether the donor site is also making some contribution.
Figure 7.
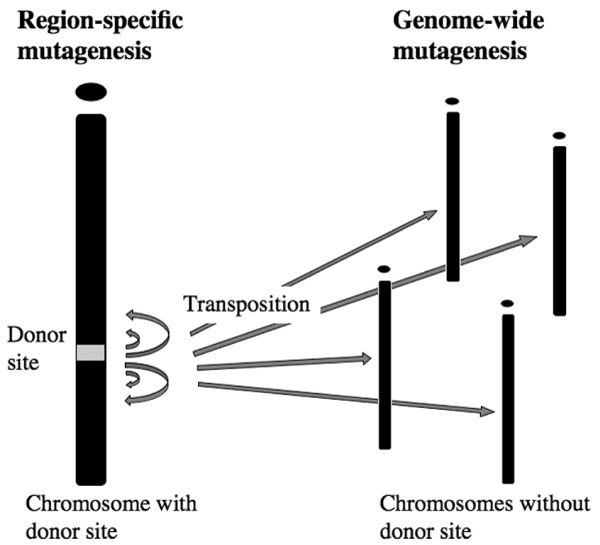
Two possible strategies for comprehensive mutagenesis using SB transposition in mice. SB, Sleeping Beauty.
In the second strategy, by focusing on transpositions with chromosomes that bear no donor site, we can easily avoid the donor site effect (Figure 7, right). In certain transgenic mice with a mobilized transposon vector, more than half of such transpositions occurred on chromosomes other than the original donor site [17]. After segregating the donor site from these mice, we will be able to generate mutant mice in a comprehensive manner without the donor site effect.
Conclusion
The SB transposon system is an effective tool as a forward genetics screen for tagged insertional mutagenesis in mice. There are many advantages of the SB transposon system relative to other mutagenesis approaches, which include the following: no embryonic stem cell manipulation is required; a simple breeding scheme allows generation of mutant mice; noninvasive screening and rapid identification of disrupted genes are possible by using the transposon sequences as a DNA tag; and, finally, region specific saturation mutagenesis is possible.
Currently, we and others have shown that this system works as an effective insertional mutagenesis screen in both germline and somatic cells [17,19-21]. An improvement to the SB transposon system for germline mutagenesis would be to increase the number of gene hits, in order to make the system more attractive to the forward genetics community and better suited to comprehensive mutagenesis screening. This can be done in many ways, including the following: optimize the transposon cargo cassette as transposition efficiency decreases with increasing vector size; screen for more effective hyperactive mutants of SB transposase for increasing transposition events; and, finally, introduce transposon vectors that can alter heterochromatin conformation change within the donor site to increase transposition efficiency.
Because SB transpositions may cause deleterious effects at the donor site concatemers [19], careful phenotypic analysis of mutant mice progeny is required, taking into consideration the possibility of a donor site effect. To overcome this limitation of SB transposition, donor sites containing single copies of SB transposon may be used. Better versions of the SB transposon system with high efficiency of transposition would make this a viable option [22].
Since the development of the SB transposon system, several other transposon systems have been reported to be active in mammalian cells, including Minos [23] (isolated from Drosophila hydei), piggyBac [24] (from the moth Trichoplusia), and Tol2 [25] (from the Japanese medaka fish Oryzias latipes). Recently, Balciunas and coworkers [26] reported that Tol2 transposase can efficiently transpose DNA sequence larger than 10 kilobases, suggesting its possible use for gene therapeutic and transgenic applications. Although further studies are needed to clarify the characteristics of these transposons, the diversity of the available tools will greatly facilitate future applications of tagged mutagenesis to functional analysis of the genome.
Competing interests
JT and KH have applied for a patent for generation of mutant animals using the SB transposon system.
Acknowledgments
Acknowledgements
We thank C Kokubu, K Yusa, M Kouno, K Yae, R Ikeda, ES Saito, Y Uno, T Hayakawa, S Kouno, and Y Odan for their advice and excellent support. This work was supported by grants from the New Energy and Industrial Technology Development Organization of Japan; RIKEN, The Institute of Physical and Chemical Research; and a grant-in-aid for Science Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
This article has been published as part of Genome Biology Volume 8, Supplement 1, 2007: Transposons in vertebrate functional genomics. The full contents of the supplement are available online at http://genomebiology.com/supplements/8/S1.
References
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/S0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Fischer SE, Wienholds E, Plasterk RH. Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci USA. 2001;98:6759–6764. doi: 10.1073/pnas.121569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K, Kuroiwa A, Ikawa M, Okabe M, Kondoh G, Matsuda Y, Takeda J. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc Natl Acad Sci USA. 2001;98:9191–9196. doi: 10.1073/pnas.161071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/S0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- Horie K, Yusa K, Yae K, Odajima J, Fischer SE, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol. 2003;23:9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Takeda J, Horie K. Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol Cell Biol. 2004;24:4004–4018. doi: 10.1128/MCB.24.9.4004-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Kokubu C, Yusa K, Keng VW, Horie K, Takeda J. Sleeping beauty transposase has an affinity for heterochromatin conformation. Mol Cell Biol. 2007;27:1665–1676. doi: 10.1128/MCB.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Kawaichi M, Ishida Y. Suppression of nonsense-mediated mRNA decay permits unbiased gene trapping in mouse embryonic stem cells. Nucleic Acids Res. 2005;33:e20. doi: 10.1093/nar/gni022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Haase I, Simmeth E, Gerisch G, Muller-Taubenberger A. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium. FEBS Lett. 2004;577:227–232. doi: 10.1016/j.febslet.2004.09.084. [DOI] [PubMed] [Google Scholar]
- Long JZ, Lackan CS, Hadjantonakis AK. Genetic and spectrally distinct in vivo imaging: embryonic stem cells and mice with widespread expression of a monomeric red fluorescent protein. BMC Biotechnol. 2005;5:20. doi: 10.1186/1472-6750-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Dickinson ME, Fraser SE, Papaioannou VE. Technicolour transgenics: imaging tools for functional genomics in the mouse. Nat Rev Genet. 2003;4:613–625. doi: 10.1038/nrg1126. [DOI] [PubMed] [Google Scholar]
- Keng VW, Yae K, Hayakawa T, Mizuno S, Uno Y, Yusa K, Kokubu C, Kinoshita T, Akagi K, Jenkins NA, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2:763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- Transposon Insertion Site Database - Germline http://transposon.abcc.ncifcrf.gov/germline/index.html
- Geurts AM, Collier LS, Geurts JL, Oseth LL, Bell ML, Mu D, Lucito R, Godbout SA, Green LE, Lowe SW, et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2:e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/S1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Klinakis AG, Loukeris TG, Pavlopoulos A, Savakis C. Mobility assays confirm the broad host-range activity of the Minos transposable element and validate new transformation tools. Insect Mol Biol. 2000;9:269–275. doi: 10.1046/j.1365-2583.2000.00183.x. [DOI] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166:895–899. doi: 10.1534/genetics.166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]


