Abstract
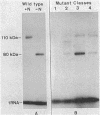
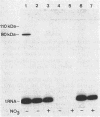
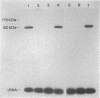
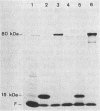
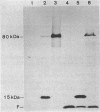
Mutants of Escherichia coli were isolated which were affected in the formation of both formate dehydrogenase N (phenazine methosulfate reducing) (FDHN) and formate dehydrogenase H (benzylviologen reducing) (FDHH). They were analyzed, together with previously characterized pleiotropic fdh mutants (fdhA, fdhB, and fdhC), for their ability to incorporate selenium into the selenopolypeptide subunits of FDHN and FDHH. Eight of the isolated strains, along with the fdhA and fdhC mutants, maintained the ability to selenylate tRNA, but were unable to insert selenocysteine into the two selenopolypeptides. The fdhB mutant tested had lost the ability to incorporate selenium into both protein and tRNA. fdhF, which is the gene coding for the 80-kilodalton selenopolypeptide of FDHH, was expressed from the T7 promoter-polymerase system in the pleiotropic fdh mutants. A truncated polypeptide of 15 kilodaltons was formed; but no full-length (80-kilodalton) gene product was detected, indicating that translation terminates at the UGA codon directing the insertion of selenocysteine. A mutant fdhF gene in which the UGA was changed to UCA expressed the 80-kilodalton gene product exclusively. This strongly supports the notion that the pleiotropic fdh mutants analyzed possess a lesion in the gene(s) encoding the biosynthesis or the incorporation of selenocysteine. The gene complementing the defect in one of the isolated mutants was cloned from a cosmid library. Subclones were tested for complementation of other pleiotropic fdh mutants. The results revealed that the mutations in the eight isolates fell into two complementation groups, one of them containing the fdhA mutation. fdhB, fdhC, and two of the new fdh isolates do not belong to these complementation groups. A new nomenclature (sel) is proposed for pleiotropic fdh mutations affecting selenium metabolism. Four genes have been identified so far: selA and selB (at the fdhA locus), selC (previously fdhC), and selD (previously fdhB).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Wolfe R. S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl Environ Microbiol. 1976 Dec;32(6):781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E. L., Jackson C. E., Fukumoto H. T., Chang G. W. Formate dehydrogenase mutants of Salmonella typhimurium: a new medium for their isolation and new mutant classes. Mol Gen Genet. 1979;177(1):95–101. doi: 10.1007/BF00267258. [DOI] [PubMed] [Google Scholar]
- Blum P., Blaha L., Artz S. Reversion and immobilization of phage Mud1 cts (Apr lac) insertion mutations in Salmonella typhimurium. Mol Gen Genet. 1986 Feb;202(2):327–330. doi: 10.1007/BF00331659. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux M., Casse F., Pascal M. C. Isolation and phenotypes of mutants from Salmonella typhimurium defective in formate hydrogenlyase activity. J Bacteriol. 1972 May;110(2):766–768. doi: 10.1128/jb.110.2.766-768.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. C., Edwards E. S., DeMoss J. A. Resolution of distinct selenium-containing formate dehydrogenases from Escherichia coli. J Bacteriol. 1981 Mar;145(3):1317–1324. doi: 10.1128/jb.145.3.1317-1324.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreiseikelmann B., Eichenlaub R., Wackernagel W. The effect of differential methylation by Escherichia coli of plasmid DNA and phage T7 and lambda DNA on the cleavage by restriction endonuclease MboI from Moraxella bovis. Biochim Biophys Acta. 1979 May 24;562(3):418–428. doi: 10.1016/0005-2787(79)90105-9. [DOI] [PubMed] [Google Scholar]
- Haddock B. A., Mandrand-Berthelot M. A. Escherichia coli formate-to-nitrate respiratory chain: genetic analysis. Biochem Soc Trans. 1982 Dec;10(6):478–480. doi: 10.1042/bst0100478. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Lester R. L., DeMoss J. A. Effects of molybdate and selenite on formate and nitrate metabolism in Escherichia coli. J Bacteriol. 1971 Mar;105(3):1006–1014. doi: 10.1128/jb.105.3.1006-1014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons L. B., Zinder N. D. The genetic map of the filamentous bacteriophage f1. Virology. 1972 Jul;49(1):45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- PINSENT J. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the coli-aerogenes group of bacteria. Biochem J. 1954 May;57(1):10–16. doi: 10.1042/bj0570010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecher A., Zinoni F., Böck A. The seleno-polypeptide of formic dehydrogenase (formate hydrogen-lyase linked) from Escherichia coli: genetic analysis. Arch Microbiol. 1985 May;141(4):359–363. doi: 10.1007/BF00428850. [DOI] [PubMed] [Google Scholar]
- Stadtman T. C. Selenium-dependent enzymes. Annu Rev Biochem. 1980;49:93–110. doi: 10.1146/annurev.bi.49.070180.000521. [DOI] [PubMed] [Google Scholar]
- Streber W. R., Timmis K. N., Zenk M. H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987 Jul;169(7):2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Leinfelder W., Böck A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc Natl Acad Sci U S A. 1987 May;84(10):3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Stadtman T. C., Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]