Abstract
TPMT is a cytosolic enzyme that catalyzes the S-methylation of aromatic and heterocyclic sulfhydryl compounds, including medications such as mercaptopurine and thioguanine. TPMT activity exhibits autosomal codominant genetic polymorphism, and patients inheriting TPMT deficiency are at high risk of potentially fatal hematopoietic toxicity. The most prevalent mutant alleles associated with TPMT deficiency in humans have been cloned and characterized (TPMT∗2 and TPMT∗3A), but the mechanisms for loss of catalytic activity have not been elucidated. In the present study, we established that erythrocyte TPMT activity was significantly related to the amount of TPMT protein on Western blots of erythrocytes from patients with TPMT activities of 0.4-23 units/ml pRBC (rs = 0.99; P < 0.001). Similarly, heterologous expression of wild-type (TPMT∗1) and mutant (TPMT∗2 and TPMT∗3A) human cDNAs in yeast and COS-1 cells demonstrated comparable levels of TPMT mRNA but significantly lower TPMT protein with the mutant cDNAs. Rates of protein synthesis were comparable for wild-type and mutant proteins expressed in yeast and with in vitro translation in rabbit reticulocyte lysates. In contrast, pulse–chase experiments revealed significantly shorter degradation half-lives for TPMT∗2 and TPMT∗3A (∼0.25 hr) compared with wild-type TPMT∗1 (18 hr). The degradation of mutant proteins was impaired by ATP depletion and in yeast with mutant proteasomes (pre-1 strain) but unaffected by the lysosomal inhibitor chloroquine. These studies establish enhanced degradation of TPMT proteins encoded by TPMT∗2 and TPMT∗3A as mechanisms for lower TPMT protein and catalytic activity inherited by the predominant mutant alleles at the human TPMT locus.
TPMT (S-adenosyl l-methionine:thiopurine S-methyltransferase; EC 2.1.1.67) is a cytosolic protein that catalyzes the S-methylation of mercaptopurine, mercaptopurine formed from the prodrug azathioprine, 6-thioguanine, and other aromatic and heterocyclic sulfhydryl compounds. TPMT activity exhibits codominant polymorphism in Caucasian and African-American populations (1, 2), with about 10% having intermediate activity due to heterozygosity at the TPMT locus and approximately 1 in 300 inheriting TPMT deficiency. Decreased S-methylation in TPMT-deficient and heterozygous patients leads to more extensive accumulation of thioguanine nucleotides (3), the principal cytotoxic metabolites (4), thereby producing greater toxicity. Patients inheriting TPMT deficiency have a pronounced risk of potentially life-threatening hematopoietic toxicity when treated with conventional doses of these medications (5–8), and patients with heterozygosity at the TPMT locus have lower activity and an intermediate risk of toxicity.
The inherited polymorphism of TPMT activity in humans has now been elucidated at the molecular level with the identification of inactivating mutations in the human TPMT gene (9, 10). The initial molecular defect to be discovered was a single G238 → C transversion mutation leading to an amino acid substitution (Ala → Pro) at codon 80 (9). Heterologous expression of this mutant allele (TPMT∗2) in yeast demonstrated TPMT mRNA levels comparable to wild type (TPMT∗1), but approximately a 100-fold reduction in S-methylation activity. The second and more prevalent mutant allele (TPMT∗3A) contains two nucleotide transition mutations (G460 → A and A719 → G) in the open reading frame, leading to amino acid substitutions Ala-154 → Thr and Tyr-240 → Cys (10, 11). Heterologous expression of the TPMT∗3A cDNA in yeast produced TPMT mRNA levels comparable to wild type, but TPMT protein levels were about 400-fold lower, with no detectable catalytic activity. However, the post-transcriptional mechanism for loss of TPMT function has not been elucidated for either TPMT∗2 or TPMT∗3A.
If the mechanism for TPMT deficiency in humans results in lower TPMT protein levels, instead of a modification in the active site(s) for metabolism of thiopurine medications, then these patients will have reduced metabolism of all TPMT substrates, including endogenous or environmental substrate(s) that have not yet been identified. The current studies were, therefore, undertaken to elucidate the mechanism(s) for loss of TPMT activity in cells expressing proteins encoded by human TPMT∗2 and TPMT∗3 alleles.
METHODS
Human Subjects.
After informed consent was obtained from the patient, parent, or guardian, blood was obtain for the measurement of TPMT activity in erythrocytes (RBCs) and for preparation of RBC lysates for Western blots. These studies were performed in unrelated individuals selected on the basis of their TPMT phenotypes: three patients known to have TPMT deficiency (4, 9, 10), four with a heterozygous TPMT phenotype (5–10 units/ml pRBC), and four with homozygous wild-type TPMT activity (>10 units/ml pRBC), by using published criteria for phenotype assignment (1). TPMT genotypes of TPMT-deficient patients were determined by cDNA cloning and sequencing, as described (9, 10).
Expression and Western Blot of TPMT Proteins.
Human TPMT cDNAs were cloned and expressed in yeast as described (9, 10). Subsequently, the coding region of TPMT cDNA was amplified by PCR using previously constructed plasmids containing TPMT∗1, TPMT∗2, or TPMT∗3A cDNA as templates, with addition of restriction sites at each end and change of thymidine to cytidine at position −1, as described (12).The primers used were the sense primer 5′-cggaattcAAACcATGGATGGTACAAGAACTTCACTTG-3′ and the antisense primers 5′-cggatccTTACTTTTCTGTAAGTAGATATAACTTTTC-3′ or 5′-cggatccTTACTTTTCTGTAAGTAGACATAACTTTTC-3′, depending on the template used. PCR products were digested with EcoRI and BamHI and then ligated with similarly digested pGEM-7Zf(+) vector (Promega). The resultant clones were subcloned into pSP64 Poly(A) vector (Promega) for in vitro translation through SalI/XhoI and BamHI restriction sites. The in vitro translation reaction was performed with TNT-Coupled Reticulocyte Lysate Systems (Promega) with the addition of 1.5 μg of plasmid DNA. TPMT cDNA inserts in pSP64 Poly(A) vector were then subcloned into pcDNA3.1(+) vector (Invitrogen) through HindIII and BamHI sites for TPMT expression in COS-1 (monkey kidney) cells transfected by the calcium phosphate method (13). Nucleotide structures of all cDNA inserts in these plasmids were confirmed by sequencing. Enzymatically active 6×His-tagged TPMT was expressed in Escherichia coli M15[pREP4] driven by a cDNA fragment containing the TPMT ORF (9) inserted into pQE-30 vector (Qiagen) and subsequently purified according to manufacturer’s instructions. Glutathione S-transferase–TPMT fusion protein was generated and purified as described (10); this fusion protein was digested with thrombin to release C-terminal TPMT and used for Western blot analysis. Expressed human TPMT proteins and TPMT in human RBC lysates and liver cytosol were analyzed by SDS/PAGE (16 cm × 16 cm; 12.5% gel) and immunodetected with TPMT-specific antibody as described (10).
Pulse–Chase Analysis.
Pulse–chase experiments were performed as described (14–16), modified as follows. Saccharomyces cerevisiae cells of the strain 2805 transformed with plasmids containing wild-type or mutant human TPMT cDNAs (10) were grown at 30°C to A600 ≈ 1.5 in 2% galactose, 0.67% yeast nitrogen base (Difco) without amino acids, adenine (20 μg/ml), and a mixture of amino acids lacking methionine or cysteine. Four mutant TPMT cDNAs were studied: TPMT∗2 with the G238C transversion (9), TPMT∗3A with both the G460A and A719G transition (10), TPMT∗3B with only the G460A mutation, and TPMT∗3C with only the A719G mutation. These cDNAs were cloned and expressed in yeast as described (9, 10). For pulse-labeling proteins, [35S]methionine (Tran35S-label, ICN) at 20 μCi/ml was added to cell cultures for 15 min. Cells were then washed and resuspended in a glucose-containing medium with amino acids including unlabeled methionine (2 mg/ml) and cysteine (0.5 mg/ml) and chased in the same medium for the indicated durations. To determine the effect of chloroquine on protein degradation, 0.2 mM chloroquine was included in the medium throughout the pulse–chase experiment. To determine the effect of ATP depletion, 2-deoxyglucose (20 mM) and 2,4-dinitrophenol (0.2 mM) were added to the medium after pulse labeling. For each experiment, 1 ml of cell culture was harvested at each time point by centrifugation, and the cell pellet was resuspended in 200 μl of 2% SDS/10 mM dithiothreitol/10 mM Tris⋅HCl, pH 7.5, and heated in a boiling-water bath for 3 min. After centrifugation, the supernatant was diluted in buffer A (1% Triton X-100/0.15 M NaCl/1 mM EDTA/0.5 mM phenylmethylsulfonyl fluoride/50 mM Tris⋅HCl, pH 7.5) and centrifuged in a Millipore Ultrafree-15 filter unit with Biomax-10 membrane, followed by dilution in buffer B (buffer A plus 10% glycerol) and centrifugation in the same unit until the volume reached 0.5–1 ml. This final filtrate was then used immediately for immunoprecipitation or stored at −70°C. The same procedure was applied for the yeast strains YWO71 (with wild-type proteasome) and YWO74 (with mutant proteasome), except that exponentially growing cells were used as described (17).
Immunoprecipitation.
The radioactivity of trichloroacetic acid-precipitable 35S-labeled protein was determined in the final filtrate. Portions of the filtrate containing equal amounts of the total acid-precipitable 35S were processed for immunoprecipitation, with a polyclonal antibody to human TPMT (10). This antibody detected TPMT∗1, TPMT∗2, TPMT∗3A, TPMT∗3B, and TPMT∗3C with similar intensity on Western blot after in vitro translation, demonstrating that this polyclonal antibody recognizes both wild-type and mutant TPMT proteins (data not shown). Antigen–antibody incubation proceeded on ice for 2 hr to overnight, protein A-Sepharose (Sigma) was then added, and the suspension was incubated on ice for an hour with vortex mixing every 3–5 min. The protein A-Sepharose pellets were washed three times with RIPA buffer (0.1% SDS/1% Triton X-100/1% sodium deoxycholate/0.15 M NaCl/1 mM EDTA/0.25 mM phenylmethylsulfonyl fluoride/50 mM Tris⋅HCl, pH 7.5), once with RIPA plus 0.5 M NaCl, and a final wash with 50 mM Tris⋅HCl (pH 7.5). The washed protein A pellets were then prepared for electrophoresis by the addition of 2× SDS/PAGE sample buffer. Electrophoresis was on 15% SDS/PAGE gels. Gels were fixed and treated with 1 M sodium salicylate; [35S]TPMT was quantitated with a PhosphorImager (Molecular Dynamics) and autoradiography was performed thereafter.
TPMT Synthesis Rate.
TPMT synthesis experiments were performed in yeast as described above, with the following modifications. The synthesis of TPMT protein in yeast was measured by pulse labeling in the galactose-containing medium for 5–20 min. After electrophoresis, gel slices corresponding to the area around the expected mobility of the protein were digested overnight in Solvable (Packard) and radioactivity of 35S was measured with the addition of scintillation fluid. The molar amount of TPMT was calculated by using the specific activity of [35S]methionine, based on four molecules of methionine in each TPMT protein and a counting efficiency of 95%. Total protein in yeast lysates was measured (18) and used to normalize the amount of TPMT synthesized. Translational efficiency was also measured in vitro in rabbit reticulocyte lysates by taking aliquots of the translated product during the linear phase of the reaction (5, 10, and 20 min) at 30°C and analyzing by SDS/PAGE and by PhosphorImager (Molecular Dynamics).
Intrinsic Stability.
To assess intrinsic stability of TPMT activity, 5–20 μl of cytosol from yeast expressing recombinant human TPMT∗1, TPMT∗3B, or TPMT∗3C were preincubated (37°C) in 250 μl of 0.1 M Tris⋅HCl (pH 7.5) buffer with or without 1 mM S-adenosylmethionine (SAM), for up to 4 hr. An aliquot of preincubation mixture was then assayed for TPMT activity with 2 mM 6-mercaptopurine and 1 mM SAM as substrates. The reaction was allowed to proceed for 10 min at 37°C, and methylmercaptopurine was measured, as described (10). The same assay mixture with yeast cytosol expressing vector alone served as the blank, and the background values were subtracted from all values obtained. There was never a sufficient amount of catalytically active TPMT protein to permit these studies with either TPMT∗2 or TPMT∗3A.
Modeling and Statistics.
A one-compartment first-order model was fit to the amount of labeled TPMT∗1, TPMT∗3A, TPMT∗3B, and TPMT∗3C, for up to 24 hr after pulse labeling of these proteins in yeast. A two-compartment model was used for fitting TPMT∗2 amount, since biphasic degradation was observed. Weighted least squares as implemented in adapt ii software (19) was used to fit the model to the data and estimate the degradation rate constants and degradation half-lives for each protein. Synthesis rates were estimated by fitting a one- or two-compartment first-order model to the amount of pulse-labeled TPMT protein from 5 to 20 min (see above) with degradation rates fixed at the rate measured from pulse–chase experiments with each protein. The 95% confidence intervals for the parameter estimates calculated in adapt ii software were used to determine whether TPMT proteins half-lives differed significantly. The relation between TPMT activity and TPMT protein levels in RBC lysates was assessed by the Spearman rank correlation coefficient (rS).
RESULTS
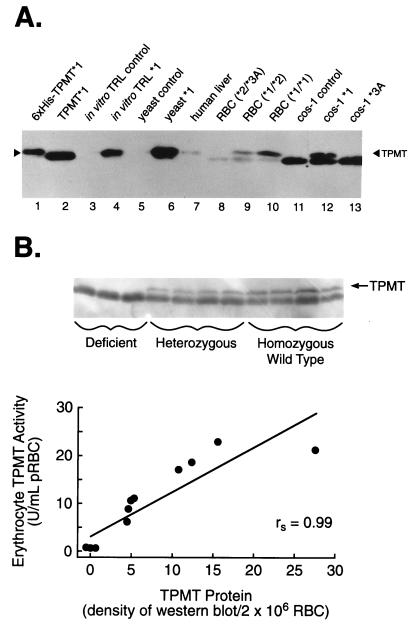
Western blots of human liver cytosol, human RBC lysates (except TPMT-deficient patients), and lysates from yeast or COS-1 cells expressing recombinant human wild-type TPMT∗1, revealed a band of approximately 32.5 kDa that was detected by the polyclonal TPMT antibody and migrated with in vitro-translated TPMT∗1 and purified TPMT (Fig. 1A). No TPMT protein was detected in controls using plasmids without TPMT cDNA inserts (Fig. 1A, lanes 3, 5, and 11). As expected, slower mobility was observed with 6×His-tagged TPMT, due to six additional histidine residues (Fig. 1A, lane 1). Densitometric quantitation of TPMT bands on Western blots revealed a significant correlation (rs = 0.99; P < 0.001) between TPMT activity and TPMT protein levels in RBC lysates from patients with TPMT activities ranging from <0.4 to 23 units/ml pRBC (Fig. 1B). Patients with two mutant alleles (TPMT∗2 and/or TPMT∗3A), were deficient in TPMT activity and had no detectable TPMT protein, whereas patients with heterozygous phenotypes (activity 5–10 units/ml pRBC) had intermediate levels of TPMT protein, and patients with homozygous wild-type phenotypes (>10 units/ml pRBC) had the highest level of TPMT protein.
Figure 1.
(A) Western blot of purified histidine-tagged human TPMT∗1 protein (0.2 ng, lane 1); TPMT∗1 generated from thrombin-treated purified glutathione S-transferase–TPMT fusion protein (2 ng, lane 2); in vitro-translated control (vector alone without TPMT cDNA insert) in rabbit reticulocyte lysate (RRL; 2 μl of 1:10 dilution, lane 3); in vitro-translated human TPMT∗1 in RRL (2 μl of 1:10 dilution, lane 4); yeast cytosol expressing vector alone (45 ng, lane 5); yeast cytosol expressing human TPMT∗1 (45 ng, lane 6); human liver cytosol (5 μg, lane 7); RBC lysates equivalent to 2 × 106 cells from patients with homozygous deficient (∗2/∗3A; 64.3 μg, lane 8), heterozygous (∗1/∗2; 87.8 μg, lane 9), or homozygous wild-type (∗1/∗1; 78.2 μg, lane 10) TPMT genotypes; COS-1 cells expressing vector alone (3.65 μg, lane 11); COS-1 cells expressing TPMT∗1 (5.01 μg, lane 12); and COS-1 cell lysate expressing TPMT∗3A (4.7 μg, lane 13). (B) Western blot of TPMT protein in RBC lysates and correlation of erythrocyte TPMT activity and protein in 11 individuals with different TPMT phenotypes. The Spearman rank order coefficient (rs) was 0.99 (P < 0.001).
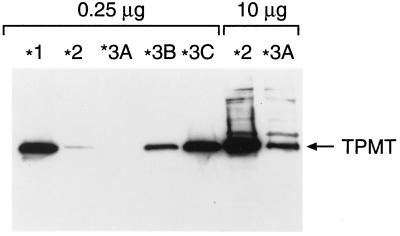
Similar to patients with different TPMT genotypes, marked differences in TPMT protein levels were observed when wild-type and mutant TPMT cDNAs were expressed in yeast (Fig. 2); with the levels of TPMT∗2 and TPMT∗3A protein about 20-fold and more than 200-fold less than TPMT∗1, respectively. Yeast expressing the TPMT∗3B cDNA with only the G460A had 4-fold lower TPMT protein, and TPMT protein levels in yeast expressing TPMT∗3C (only A719G) were similar to wild type. Comparable results were obtained when these mutant cDNAs were expressed in COS-1 cells (for TPMT∗3A, see Fig. 1A, lane 13; data not shown for other mutants). TPMT mRNA levels were comparable in yeast expressing wild-type or any of the four mutant cDNAs (9, 10), indicating that these mutations did not affect transcription under these conditions, implicating post-transcriptional mechanisms as the basis for TPMT deficiency.
Figure 2.
Western blot of recombinant human TPMT protein in yeast cytosol expressing wild-type (∗1) and mutant TPMT cDNAs (∗2, ∗3A, ∗3B, and ∗3C) after a 24-hr culture in galactose-containing medium. The amount of total protein loaded in each lane is indicated at the top.
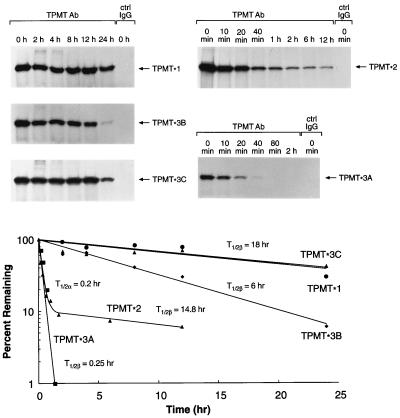
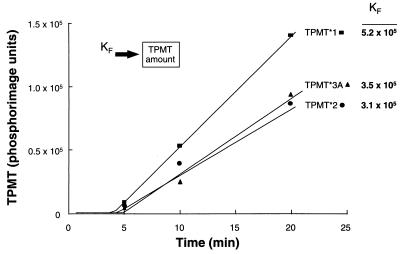
By using pulse–chase techniques, significant differences were found in half-lives of wild-type and mutant TPMT proteins in yeast, with a degradation half-life of 18 hr for TPMT∗1 and TPMT∗3C, 6 hr for TPMT∗3B, 0.2 hr (α) and 14.8 hr (β) for TPMT∗2, and 0.25 hr for TPMT∗3A (Fig. 3). In contrast, synthesis rates were similar for the wild-type and mutant TPMT proteins expressed in yeast (Table 1). Similar translational rates were also observed when TPMT∗1, TPMT∗2, and TPMT∗3A were translated in vitro in rabbit reticulocyte lysates (Fig. 4). In yeast expressing wild-type or mutant TPMT cDNAs, the synthesis and degradation rates for each protein were in good agreement with relative levels of TPMT protein measured after 24 hr of culture (Fig. 2).
Figure 3.
Degradation kinetics of recombinant human TPMT proteins in yeast expressing wild-type (TPMT∗1) and mutant TPMT cDNAs. Yeast cells were pulse-labeled with [35S]methionine (Tran35S-label) and chased for the duration of time indicated. 35S-labeled TPMT was immunoprecipitated with TPMT antibody or control rabbit IgG and electrophoresed on SDS/PAGE gels. (Upper) Pulse–chase experiments for wild-type (TPMT∗1) and mutant proteins (TPMT∗2, ∗3A, ∗3B, and ∗3C). (Lower) TPMT degradation from PhosphorImager quantitation of 35S-labeled proteins.
Table 1.
Half-lives and synthesis rates of wild-type and mutant TPMT proteins in yeast
| Parameter | TPMT∗1 | TPMT∗2 | TMPT∗3A | TMPT∗3B | TPMT∗3C |
|---|---|---|---|---|---|
| Degradation t1/2, hr | 18 (0.4) | 0.2α (43), | 0.25 (6)* | 6.1 (0.3)* | 18 (0.3) |
| 14.8β (1)* | |||||
| Formation rate fmol per mg per hr | 335 (8) | 409 (9) | 268 (14) | 349 (8) | 220 (12) |
Data in parentheses are coefficient of variation percent.
P < 0.05, when compared to TPMT ∗ 1.
Figure 4.
In vitro translation of TPMT∗1, ∗2, and ∗3A in rabbit reticulocyte lysate. Formation rates (Kf) were estimated by fitting the model (Inset) to the amount of 35S-labeled TPMT protein versus time data, when no degradation of TPMT proteins in this in vitro translation system is assumed.
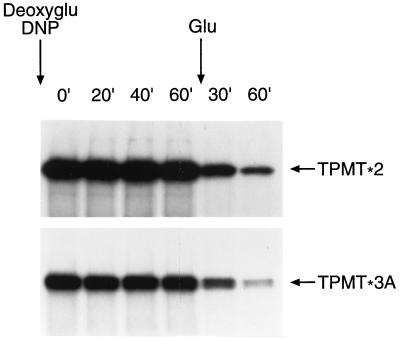
Two major proteolytic pathways are responsible for the degradation of proteins in eukaryotic cells; an ATP-dependent proteasomal pathway and an ATP-independent lysosomal pathway (20, 21). To assess the requirement of ATP in the degradation of mutant TPMT proteins, 2-deoxyglucose and 2,4-dinitrophenol were used to inhibit ATP production and thus deplete intracellular ATP in yeast expressing mutant TPMT proteins (22, 23). As depicted in Fig. 5, the proteolysis of TPMT∗2 and TPMT∗3A was inhibited in the ATP-depleting medium and rapidly restored by resuspending cells in glucose-containing medium.
Figure 5.
Degradation of 35S-labeled recombinant human TPMT∗2 and TPMT∗3A in yeast, chased first in ATP-depleting medium containing 20 mM 2-deoxyglucose (deoxyglu) and 0.2 mM 2,4-dinitrophenol (DNP) for the time (min) indicated above each lane and then in glucose-containing medium.
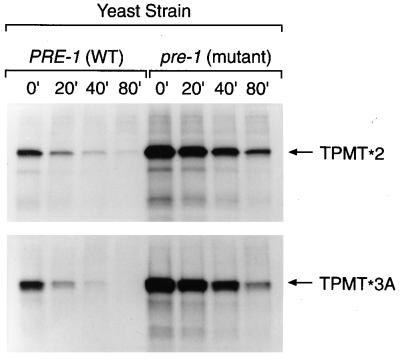
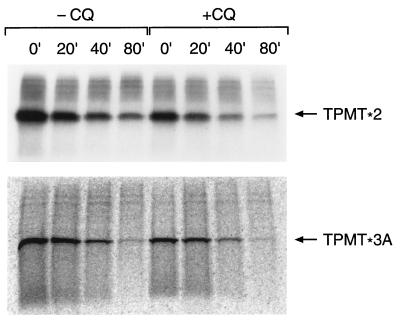
Because the ATP-dependent nature of TPMT degradation suggested a proteasomal mechanism, a proteasome-mutant yeast strain (pre-1) was used to further characterize proteasome-mediated degradation of TPMT. As depicted in Fig. 6, both TPMT∗2 and TPMT∗3A were degraded more slowly in the pre-1 strain compared with the yeast with wild-type proteasomes. With equal amounts of radiolabeled proteins used in the immunoprecipitation, the accumulation of labeled TPMT protein in the pre-1 strain at time zero is consistent with impaired TPMT degradation during the time of pulse labeling. In contrast, there was no significant difference in degradation kinetics when cells were incubated in the presence or absence of chloroquine (Fig. 7), at a concentration (0.2 mM) previously shown to inhibit lysosomal metabolism in yeast (24).
Figure 6.
Degradation of 35S-labeled TPMT∗2 and TPMT∗3A expressed in YWO71 (PRE-1 with wild-type proteasome) and YWO74 (pre-1 with mutant proteasome), analyzed by pulse–chase experiments. Numbers (min) above each lane indicate the duration of the chase period. An equal amount of radiolabeled protein was loaded in each lane.
Figure 7.
Degradation of 35S-labeled recombinant human TPMT∗2 and TPMT∗3A in yeast, pulse-chased in the presence (+CQ) or absence (−CQ) of 0.2 mM chloroquine. Numbers (min) above each lane indicate the duration of the chase period. An equal amount of radiolabeled protein was loaded in each lane. (Upper) Autoradiograph. (Lower) Phosphorimage.
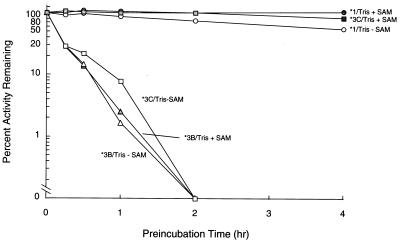
As depicted in Fig. 8, there was rapid loss of catalytic activity for TPMT∗3B and TPMT∗3C when incubated at 37°C in Tris buffer at pH 7.5, whereas wild-type TPMT activity was much more stable under these conditions (TPMT∗2 and TPMT∗3A were too rapidly degraded in the heterologous expression system to permit analysis of intrinsic stability). The addition of cosubstrate SAM (1 mM) resulted in partial stabilization of catalytic activity for TPMT∗3C (Fig. 8) but not for TPMT∗3B protein. Under all conditions for assessing intrinsic stability, there was no loss of immunodetectable protein over the time periods studied (data not shown).
Figure 8.
Intrinsic stability of TPMT proteins encoded by wild-type (TPMT∗1) and mutant TPMT cDNAs (TPMT∗3B and TPMT∗3C). After incubation in Tris buffer at pH 7.5 with (filled symbols) or without (open symbols) 1 mM SAM, S-methylation of mercaptopurine was measured. Catalytic activity is expressed as a percentage of the initial activity at time zero (no preincubation).
DISCUSSION
The polymorphism of TPMT activity was discovered more than 15 years ago (1), yet the genetic basis for this inherited trait was not known until recently (9, 10). Although these studies have established that TPMT∗2 and TPMT∗3A are mutant alleles associated with inheritance of TPMT deficiency (9, 10), the mechanism for loss of TPMT activity has not been previously elucidated.
The current study has established a significant relation between RBC TPMT activity and the level of TPMT protein in RBC lysates from patients with differing TPMT phenotypes (Fig. 1B). The presence of comparable TPMT mRNA levels in patients with TPMT deficiency and those with heterozygous or homozygous wild-type phenotypes (9, 10) and the absence of TPMT protein in TPMT-deficient patients indicate a post-transcriptional mechanism for TPMT deficiency. This is consistent with results in our heterologous expression system, wherein expression of wild-type and mutant TPMT cDNAs revealed comparable TPMT mRNA levels (9, 10), but yeast expressing either TPMT∗2 or TPMT∗3A had significantly lower TPMT protein levels compared with yeast expressing the wild-type cDNA (Fig. 2). This difference in TPMT protein was also evident when wild-type and mutant TPMT was heterologously expressed in mammalian COS-1 cells (Fig. 1A). Thus, both humans and yeast expressing TPMT∗2 and TPMT∗3A have markedly lower TPMT protein levels and catalytic activity, despite TPMT mRNA levels comparable to wild type. A second, invariant, lower molecular weight band was also observed in human RBC and monkey kidney COS-1 cell lysates. Direct sequencing of this second band in human RBC lysates indicates that it is the hemoglobin β-chain (data not shown), which may react with the antibody due to its abundance in RBC lysates. The second lower molecular weight band in COS-1 cells may reflect endogenous TPMT in these cells.
Pulse–chase and in vitro translation experiments established that lower TPMT protein levels in yeast expressing mutant human TPMT∗2 or TPMT∗3A cDNAs are due to differences in degradation rates of mutant proteins (Fig. 3) and not differences in synthesis rates (Fig. 2 and Table 1). These results indicate that the G238C, G460A, and A719G point mutations in TPMT cDNAs have little or no effect on transcription or translational efficiency but can have a significant effect on protein degradation. The G238C mutation (Ala-80 → Pro) had the greatest effect on protein degradation (t½α = 0.2 hr), and the G460A (Ala-154 → Tyr) mutation had a modest effect (t½ = 6 hr) and the A719G (Tyr-240 → Cys) mutation alone had no effect on TPMT protein degradation in yeast. Models of the TPMT secondary structure using the Garnier algorithm (25), indicate that the Ala-80 → Pro substitution produces an additional turn in the secondary structure of the TPMT protein, and the G460A mutation (Ala-154 → Tyr) produces an additional turn and a loss of one β-sheet, but no changes are predicted for the A719G mutation. Thus, the G238C and G460A mutations may result in more disruption of the tertiary structure than the A719G mutation, consistent with more rapid degradation of TPMT∗2 and TPMT∗3B. Moreover, when the G460A and A719G mutations are present together, as is the case for the most prevalent mutant allele in humans (TPMT∗3A), there was a dramatic increase in the rate of protein degradation (Fig. 3), suggesting that there is a greater effect on the tertiary structure of TPMT when both mutations are present, thus making it more susceptible to proteolysis.
Significant differences were also observed in the intrinsic stability of mutant proteins. When incubated at 37°C in Tris buffer at pH 7.5, proteins encoded by TPMT∗3B or TPMT∗3C (with either the G460A or A719G mutation alone, respectively) completely lost catalytic activity within 4 hr, without evidence of protein degradation, whereas proteins encoded by TPMT∗1 retained ∼80% of its catalytic activity, demonstrating that intrinsic stability of proteins encoded by TPMT∗3B and TPMT∗3C are significantly different from wild type. The addition of high concentrations of SAM (1 mM) partially stabilized TPMT∗3C catalytic activity under these conditions (Fig. 8), which could be explained by stabilization of the tertiary structure in the presence of substrate, as has been reported for other enzymes (26). It remains to be determined whether TPMT∗3C is stabilized by endogenous substrate(s) in yeast, thereby preventing its rapid degradation. However, it is clear from the present work that enhanced protein degradation is a primary mechanism for loss of TPMT activity with TPMT∗2 and TPMT∗3A, which represent >80% of mutant TPMT alleles in Caucasians (27).
The present work further indicates that the mechanism for proteolysis of mutant TPMT is via an ATP-dependent proteasome-mediated pathway, a mechanism known to degrade a number of intracellular proteins, including those with misfolded structures due to mutations (28, 29). Degradation of mutant TPMT proteins was not affected by the lysosomal inhibitor chloroquine (30, 31), but degradation was impaired in the mutant pre-1 yeast (15) (Figs. 7 and 8). It should be noted that some degradation of TPMT∗2 and TPMT∗3A proteins occurred in the pre-1 proteasome mutants, which lacks the “chymotrypsin-like” activity of the multi-catalytic protein complex, but retains trypsin-like and peptidyl-glutamylpeptide hydrolyzing activities (32). Thus, one of the remaining proteolytic activities in pre-1 proteasomes may also mediate degradation of mutant TPMT proteins or an additional nonlysosomal pathway may contribute to the degradation of these proteins.
It is clear from the current studies that TPMT deficiency inherited by TPMT∗2 and TPMT∗3A, the most prevalent mutant TPMT alleles in humans, is associated with lower cellular levels of TPMT protein and that the proteins encoded by these mutant alleles are degraded more rapidly by an ATP-dependent proteasome-mediated pathway. Although enhanced proteolysis of mutant proteins is not uncommon, there are many examples of mutations that lead to loss or gain of function without enhanced degradation [e.g., CFTR (33), steroid 21-hydroxylase (34), and p53 (35)]. An additional implication of enhanced proteolysis of mutant TPMT is that the absence of TPMT protein in those inheriting TPMT deficiency means these individuals will have decreased metabolism of all TPMT substrates, including as yet undiscovered environmental and endogenous substrate(s) for this polymorphic enzyme.
Acknowledgments
We thank Amy Atkinson, YaQin Chu, Natasha Lenchik, and Jackie Shireman, for their excellent technical assistance. Special thanks go to Dr. John Schuetz, Dr. Sandra D’Azzo, and Dr. Daxi Sun for their insightful discussions and advice and to the patients and parents who participated in this study. We are also most grateful to Drs. Petra Hubbe and A. Jentsch who provided the Pre-1 mutant and PRE1 wild-type yeast, and Dr. Clayton Naeve of the St. Jude Biotechnology Shared Resource. This work was supported in part by National Institutes of Health grants (R37 CA36401, Leukemia Program Project Grant CA20180, and Cancer Center CORE Grant CA21765), by a Center of Excellence grant from the State of Tennessee, and by American Lebanese Syrian Associated Charities.
ABBREVIATIONS
- TPMT
thiopurine S-methyltransferase
- RBC
red blood cell
- SAM
S-adenosylmethionine
- pRBC
packed RBC
References
- 1.Weinshilboum R M, Sladek S L. Am J Hum Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- 2.McLeod H L, Lin J-S, Pui C-H, Scott E P, Evans W E. Clin Pharmacol Ther. 1994;55:15–20. doi: 10.1038/clpt.1994.4. [DOI] [PubMed] [Google Scholar]
- 3.Lennard L, Van Loon J A, Weinshilboum R M. Clin Pharmacol Ther. 1989;46:149–154. doi: 10.1038/clpt.1989.119. [DOI] [PubMed] [Google Scholar]
- 4.Elion G B. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- 5.Evans W E, Horner M H, Chu Y Q, Kalwinsky D, Roberts W M. J Pediatr. 1991;119:985–989. doi: 10.1016/s0022-3476(05)83063-x. [DOI] [PubMed] [Google Scholar]
- 6.Lennard L, Gibson B E S, Nicole T, Lilleyman J S. Arch Dis Child. 1993;69:577–579. doi: 10.1136/adc.69.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schutz E, Gummert J, Mohr F, Oellerich M. Lancet. 1993;341:436. doi: 10.1016/0140-6736(93)93028-y. (lett.). [DOI] [PubMed] [Google Scholar]
- 8.McLeod H L, Miller D R, Evans W E. Lancet. 1993;341:1151. doi: 10.1016/0140-6736(93)93168-z. (lett.). [DOI] [PubMed] [Google Scholar]
- 9.Krynetski E Y, Schuetz J D, Galpin A J, Pui C-H, Relling M V, Evans W E. Proc Natl Acad Sci USA. 1995;92:949–953. doi: 10.1073/pnas.92.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai H L, Krynetski E Y, Yates C R, Loennechen T, Fessing M, Krynetskaia N F, Evans W E. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 11.Szumlanski C, Otterness D, Her C, Lee D, Brandriff B, Kelsell D, Spurr N, Lennard L, Wieben E, Weinshilboum R. DNA Cell Biol. 1996;15:17–30. doi: 10.1089/dna.1996.15.17. [DOI] [PubMed] [Google Scholar]
- 12.Honchel R, Aksoy I A, Szumlanski C, Wood T C, Otterness D M, Wieben E D, Weinshilboum R M. Mol Pharmacol. 1993;43:878–887. [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Vol. 3. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 16.17–16.40. [Google Scholar]
- 14.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 15.Wittenberg C, Reed S I. Cell. 1988;54:1061–1072. doi: 10.1016/0092-8674(88)90121-3. [DOI] [PubMed] [Google Scholar]
- 16.McCracken A A, Kruse K B. Mol Biol Cell. 1993;4:729–736. doi: 10.1091/mbc.4.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seufert W, Jentsch S. EMBO J. 1992;11:3077–3080. doi: 10.1002/j.1460-2075.1992.tb05379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henkel A W, Bieger S C. Anal Biochem. 1994;223:329–331. doi: 10.1006/abio.1994.1595. [DOI] [PubMed] [Google Scholar]
- 19.D’Argenio D Z, Schumitzky A. Comput Programs Biomed. 1979;9:115–134. doi: 10.1016/0010-468x(79)90025-4. [DOI] [PubMed] [Google Scholar]
- 20.Ciechanover A, Schwartz A L. FASEB J. 1994;8:182–191. doi: 10.1096/fasebj.8.2.8119489. [DOI] [PubMed] [Google Scholar]
- 21.Olson T S, Dice J F. Curr Opin Cell Biol. 1989;1:1194–1200. doi: 10.1016/s0955-0674(89)80071-7. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg-Hasson Y, Bercovich Z, Ciechanover A, Kahana C. Eur J Biochem. 1989;185:469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- 23.Shaeffer J R. J Biol Chem. 1983;10:13172–13177. [PubMed] [Google Scholar]
- 24.Finkelstein D B, Strausberg S. J Biol Chem. 1979;254:796–803. [PubMed] [Google Scholar]
- 25.Genetic Computer Group. Program Manual for the GCG Package. Madison, WI: Genetics Computer Group; 1991. , Version 7. [Google Scholar]
- 26.Johnston J A, Johnson E S, Waller P R H, Varshavsky A. J Biol Chem. 1995;270:8172–8178. doi: 10.1074/jbc.270.14.8172. [DOI] [PubMed] [Google Scholar]
- 27.Yates C R, Krynetski E Y, Loennechen T, Fessing M Y, Tai H-L, Pui C-H, Relling M V, Evans W E. Ann Int Med. 1997;126:608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 28.Rechsteiner M. Cell. 1991;66:615–618. doi: 10.1016/0092-8674(91)90104-7. [DOI] [PubMed] [Google Scholar]
- 29.Hershko A, Eytan E, Ciechanover A, Haas A L. J Biol Chem. 1982;257:13964–13970. [PubMed] [Google Scholar]
- 30.Levade T, Vidal F, Vermeersch S, Andrieu N, Gatt S, Salvayre R. Biochim Biophys Acta. 1995;1258:277–287. doi: 10.1016/0005-2760(95)00132-v. [DOI] [PubMed] [Google Scholar]
- 31.Seglen P O, Grinde B, Solheim A E. Eur J Biochem. 1979;95:215–225. doi: 10.1111/j.1432-1033.1979.tb12956.x. [DOI] [PubMed] [Google Scholar]
- 32.Heinemeyer W, Kleinschmidt J A, Saidowsky J, Escher C, Wolf D H. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welsh M J, Smith A E. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 34.Amor M, Parker K L, Globerman H, New M I, White P C. Proc Natl Acad Sci USA. 1988;85:1600–1604. doi: 10.1073/pnas.85.5.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reich N C, Oren M, Levine A J. Mol Cell Biol. 1983;312:2143–2150. doi: 10.1128/mcb.3.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]