Abstract
The effectiveness of ongoing gene therapy trials may be limited by the expression characteristics of viral and plasmid-based vectors. To enhance levels of heterologous gene expression, we have developed a safety-modified episomal expression vector that replicates extrachromosomally in human cells. This vector system employs a simian virus 40 (SV40) large T antigen mutant (107/402-T) that is deficient in binding to human tumor suppressor gene products, including p53, retinoblastoma, and p107, yet retains replication competence. These SV40-based episomes replicate to thousands of copies by 2–4 days after gene transfer in multiple types of human cell lines, with lower activity in hamster cells, and no detectable activity in dog, rat, and murine cell lines. Importantly, 107/402-T has enhanced replication activity compared with wild-type T antigen; this finding may be due, in part, to the inability of p53 and retinoblastoma to inactivate 107/402-T function. We demonstrate that the level and duration of 107/402-T expression regulates the observed episomal copy number per cell. Compared with standard plasmid constructs, episomes encoding 107/402-T yield approximately 10- to 100-fold enhanced levels of gene expression in unselected populations of transient transfectants. To determine if 107/402-T-based episomes replicate extrachromosomally in vivo, tumor explants in nude mice were directly injected with liposome/DNA complexes. Using a PCR-based assay, we demonstrate that SV40-based episomes replicate in human cells after direct in vivo gene transfer. These data suggest that safety-modified SV40-based episomes will be effective for cancer gene therapy because high level expression of therapeutic genes in transient transfectants should yield enhanced tumor elimination.
Keywords: simian virus 40 large T antigen, episomes
Limitations in the expression characteristics of viral and plasmid-based expression vectors likely curtail the effectiveness of therapeutic genes currently being tested in clinical gene therapy trials. For example, cells infected with retroviral vectors typically have only one or two integrated proviral copies per cell (1), whereas a higher copy number of transcriptional units would likely increase levels of gene expression. Similarly, vectors that persist as nonreplicating extrachromosomal elements, including adenoviral vectors and standard plasmid constructs, are subject to destruction by nucleases and can otherwise be functionally inactivated by partitioning to non-nuclear compartments. Furthermore, the copy number of extrachromosomal DNA per cell falls exponentially when replicating cell populations are the target of gene transfer (2). Refinements in the design of nonreplicating vectors, including use of cis- and trans-elements to improve levels of gene expression, do not address the significant problems of vector copy number destruction, inactivation, or dilution.
To overcome these significant limitations and thereby optimize levels of heterologous gene expression, DNA vectors can be modified to permit extrachromosomal replication in human cells. This modification can be accomplished by including replicons from DNA viruses that infect human and primate cells, including Epstein–Barr virus (3), BK virus (4, 5), and simian virus 40 (SV40) (6). Such replicons consist of two elements: (i) a viral DNA origin of replication and (ii) a viral early gene product that functions as a replication transactivator. As we have previously demonstrated for BK-virus-derived constructs (5), replicating episomal vectors have two advantages compared with standard plasmids: (i) high level gene expression due to vector amplification and (ii) maintenance of gene expression in transiently transfected cells due to efficient vertical transfer of the episome during cell division. However, a significant obstacle to the development of this class of vectors is the transformation properties associated with suitable viral early genes that possess replication transactivator function. For example, the Epstein–Barr virus replication transactivator, EBNA-1, is tumorigenic in transgenic mice (7). In addition, papovavirus early gene products, including the large T antigens from BK virus and SV40, have transformation properties thought to be primarily mediated by binding to host tumor suppressor gene products, including p53, retinoblastoma (RB), and RB-related proteins, such as p107 and p130 (8–11).
We have developed a strategy to safety-modify episomal vectors by constructing a novel SV40 large T antigen mutant that is deficient in binding human tumor suppressor gene products yet retains replication competence. We demonstrate that our SV40-based episomal vectors replicate extrachromosomally in human tumor cells in vitro, and after direct in vivo gene transfer into established tumor explants. These vectors yield high level gene expression that is maintained for at least 1 week after gene transfer. Our vector system will therefore be particularly appropriate for cancer gene therapy, in which transient, high level expression of therapeutic genes is predicted to yield efficient tumor elimination.
MATERIALS AND METHODS
Cell Lines.
Unless otherwise specified, all cell lines were obtained from American Type Culture Collection. Hep G2 cells were grown in 50% DMEM and 50% F12 media (GIBCO) supplemented with 10% heat-inactivated fetal calf serum (FCS) (HyClone); MCF-7, in RPMI 1640 medium (GIBCO) with 25 μg/ml insulin and 10% FCS; MDCK-2 and D17, in Eagle’s MEM (GIBCO) with 10% FCS; V79, in MEM-α (GIBCO) with 5% FCS; PC-12, in DMEM with 5% FCS and 10% horse serum (GIBCO); F9, in DMEM with 15% FCS; and 3T3, in DMEM with 10% calf serum (GIBCO). All other cell lines were grown in DMEM with 10% FCS. Each media preparation was supplemented with 50 μg/ml penicillin, 50 units/ml streptomycin, and 2 mM glutamine. Cells were cultured at 37°C in 5% CO2 except for PC-12 cells, which were grown at 37°C in 7% CO2.
Episomal Plasmid Construction and Reporter Vectors.
Wild-type SV40 large T antigen cDNA was isolated from plasmid pSG5-T as a 2.1-kb BamHI fragment. After XbaI linker addition, T antigen cDNA was ligated in the unique XbaI site of pRC/CMV (Invitrogen) to form pRC/CMV-T. In this vector, T antigen cDNA is transcriptionally controlled by the cytomegalovirus (CMV) immediate-early promoter. pRC/CMV contains an SV40 DNA origin; pRC/CMV-T, therefore, contains a complete SV40 replicon. In a similar fashion, pRC/CMV.107-T was constructed from pSG5-K1, which encodes a mutant T antigen substituting lysine for glutamic acid at codon 107 (12). pRC/CMV.402-T and pRC/CMV.107/402-T were constructed by substituting a 1,067-bp HpaI C-terminal fragment of T antigen from pRC/CMV-T and pRC/CMV.107-T, respectively, with the corresponding T antigen fragment from a mutant SV40 clone that encodes a point mutation which substitutes glutamic acid for aspartic acid at codon 402 (clone 402DE) (13). DNA sequence analysis confirmed in-frame ligation of the HpaI fragment and also verified the presence or absence of point mutations in codons 107 and 402 for each plasmid construct.
pRSVlacZII encodes the β-galactosidase gene transcriptionally controlled by the RSV (Rous sarcoma virus) long terminal repeat, and contains an SV40 DNA origin (14). pCMVint-lux encodes the luciferase gene regulated by the CMV immediate-early promoter, and lacks an SV40 DNA origin (15).
Immunoprecipitation Analysis.
Wild-type and mutant T antigens were translated in vitro in the presence of [35S]methionine using a reticulocyte lysate system as described by the manufacturer (Promega). Labeled T antigen (2 × 105 dpm) was added to extracts from CV-1 cells in which human RB, p107, or p53 was transiently expressed at high levels. CV-1 cells were infected with a vaccinia virus vector encoding T7 RNA polymerase, and 1 h later cells were transfected with derivatives of the pTM1 plasmid (16) containing a T7 polymerase site immediately upstream of human RB, p107, or p53 cDNA. Approximately 18 h later, cells were harvested using a lysis buffer as described (17). Immunoprecipitation analysis was performed using monoclonal antibodies to RB (clone G3–245, PharMingen), p107 (clone SD9, Oncogene Science), and p53 (clone 1801, Oncogene Science), as described (9).
Transfection.
Dishes (100 mm) of HT-1376 cells, approximately 50% confluent, were transfected using 42 μg of DNA and 120 μl of lipofectin (GIBCO) as described (5). Hep G2 cells were transfected with 14 μg of DNA per 100-mm dish using the calcium phosphate method (18). RAJI cells (2 × 106) were electroporated with 20 μg of DNA at a setting of 200 V and 750 μF. Specific transfection conditions for other cell types listed in Table 2 were optimized to achieve a transfection efficiency of at least 1% while minimizing cellular toxicities. The day after gene transfer, cells were split to maintain log phase growth for the duration of the experiment.
Table 2.
Replication activity of 107/402-T-based episomes in human and animal cell lines
| Species | Cell Line | Type | Copy number per cell* |
|---|---|---|---|
| Human | HT-1376 | Bladder | 1,400 |
| 5637 | Bladder | 100,000 | |
| MCF-7 | Breast | 8,600 | |
| T98G | Brain | 25,000 | |
| SW480 | Colon | 78 | |
| Hs68 | Fibroblast | 82 | |
| Hep G2 | Hepatoma | 25,000 | |
| NCI-H69 | Lung | 9,000 | |
| NCI-H82 | Lung | 1,200 | |
| NCI-H146 | Lung | 2,200 | |
| RAJI | Lymphoma | 7,000 | |
| Simian | CV-1 | Kidney | 11,000 |
| Dog | MDCK-2 | Kidney | <1 |
| D17 | Osteosarcoma | <1 | |
| Hamster | BHK | Kidney | <1 |
| V79 | Lung | 35 | |
| Rat | PC12 | Pheochromocytoma | <1 |
| Mouse | F9 | Embryonal carcinoma | <1 |
| 3T3 | Fibroblast | <1 |
Peak copy number was achieved between days 2 and 6.
Southern Blot Analysis of Episomal Replication.
DNA harvested from transient transfectants was evaluated for presence of extrachromosomal plasmid replication by resistance to DpnI digestion as described (5). Episomal copy number per cell was calculated based on band intensities of experimental lanes compared with a standard curve of plasmid DNA, the transient transfection efficiency, and cell DNA content. The gene transfer efficiency was experimentally determined for each cell line in Table 2 based on the percentage of blue-stained cells observed 1 day after transfection with pRSVlacZII (14).
Western Blot Analysis.
Western blot analysis of 107/402-T expression was performed using anti-T antigen monoclonal antibody 416 (Oncogene Science) and chemiluminescent substrates (Amersham) as described (17).
Luciferase and β-Galactosidase Assays.
Extracts from transient transfectants were prepared using a cell lysis buffer (Tropix, Bedford, MA), and protein was quantitated using the Bio-Rad DC method. Samples were assayed in triplicate using chemiluminescent substrates for luciferase (Promega) and β-galactosidase (Tropix) enzymes. To specifically inactivate endogenous β-galactosidase activity, cell extracts were heated to 50°C for 1 h before assaying for heterologous β-galactosidase activity (19).
In Vivo Gene Transfer.
Lightly anesthetized 4- to 6-week-old female nude mice bearing established subcutaneous HT-1376 xenografts, approximately 1 × 1 cm in size, were directly injected with 0.8 ml of liposome/DNA complexes consisting of 10 μg of plasmid DNA and 40 μl of lipofectin (GIBCO) in PBS.
PCR-Based Episomal Replication Assay.
Three days after gene transfer with pRC/CMV or pRC/CMV.107/402-T, total cellular DNA was prepared from HT-1376 tumor explants, and samples were evaluated for extrachromosomal plasmid replication using a PCR-based assay (20). Before PCR amplification, 1 μg of DNA was digested with 20 units of DpnI enzyme at 37°C overnight, and the enzyme was then inactivated by heating the sample to 65°C for 10 min. Based on the methylation-sensitivity of DpnI, this step limits generation of amplification products to newly replicated, episomal DNA. Samples were then subjected to PCR amplification using primers that generate a 1.6-kbp fragment extending from the bovine growth hormone polyadenylylation site to coding sequences in the neomycin resistance gene (REP1, 5′-GCT CGC TGA TCA GCC TCG AC-3′; REP2, 5′-CGA ACA GTT CGG CTG GCG CG-3′). After an initial denaturation for 3 min at 95°C, 32 cycles of amplification were performed using cycle times of 1 min at 95°C, 1 min at 60°C, and 3 min at 72°C. A 5-min extension step at 72°C followed the PCR amplification. Amplification products were evaluated by Southern blot analysis using a hybridization probe generated from a pair of internally nested primers (REP1B, 5′-GTG CCT TCT AGT TGC CAG CC-3′; REP2B, 5′-CGA CAA GAC CGG CTT CCA TC-3′).
Statistical Analysis.
Gene expression results are expressed as the mean ± SD. A Student’s t test was used to evaluate differences in levels of control and episome-mediated gene expression.
RESULTS
Replication-Competent, Safety-Modified SV40 Large T Antigen Mutants.
To develop an SV40-based episomal vector suitable to use for human gene therapy, we evaluated a panel of large T antigen mutants that were predicted to lack transformation properties and possibly retain replication competence. Of particular interest were the K1 mutant, which encodes a point mutation in codon 107 and lacks binding to RB protein (9, 12), and the 402DE mutant, which encodes a point mutation in codon 402 and lacks binding to p53 (13). We constructed a novel T antigen mutant, 107/402-T, that incorporates both point mutations and is, therefore, predicted to lack binding to RB and p53 (Fig. 1A). We hypothesized that 107/402-T also might retain replication competence because SV40 encoding either T antigen mutant alone forms plaques in permissive cells (12, 13).
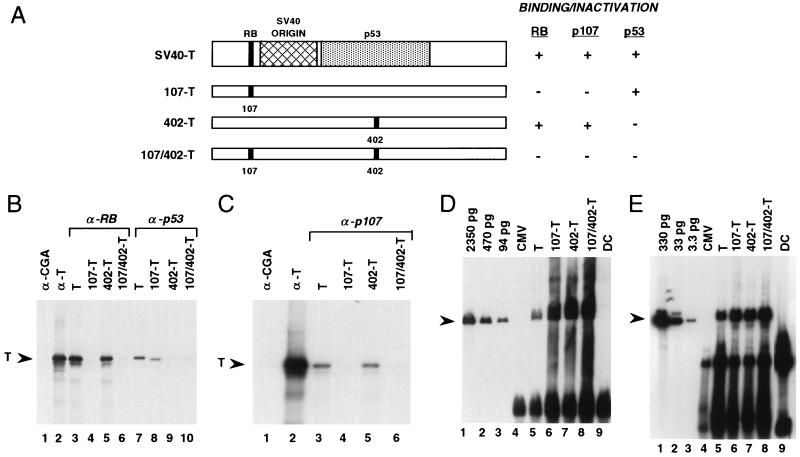
Figure 1.
107/402-T lacks binding to human tumor suppressor genes and is replication-competent. (A) Point mutations in replication-competent, safety-modified SV40 large T antigen mutants. Highlighted are domains of T antigen that bind to RB, p53, and the SV40 DNA origin. The codon 107 mutation substitutes lysine for glutamic acid, and the codon 402 mutation substitutes glutamic acid for aspartic acid (12, 13). (B and C) Coimmunoprecipitation analysis of binding of wild-type and mutant T antigens to human tumor suppressor gene products. In vitro translated T antigens (2 × 105 dpm) were mixed with CV-1 extracts overproducing human RB protein and anti-RB monoclonal antibody G3–245 (B, lanes 3–6), p53 and anti-p53 monoclonal antibody 1801 (B, lanes 7–10), and p107 and anti-p107 monoclonal antibody SD9 (C, lanes 3–6). As controls, wild-type T antigen is immunoprecipitated with either anti-chromogranin A monoclonal antibody LKH210 (lane 1) or anti-T antigen monoclonal antibody 416 (lane 2). (D and E) 107/402-T is replication-competent. Hep G2 hepatoma cells (D) were transfected with wild-type and mutant T antigen expression vectors, and total cellular DNA was harvested 2 days after transfection. DNA samples were sequentially digested with ApaI to linearize vector DNA and then with DpnI to distinguish amplified DNA from the input DNA used to transfect these cells. Because human cells lack adenine methylase activity, newly replicated DNA is resistant to digestion by DpnI. Hence the presence of unit length, linearized plasmid DNA, as indicated by the arrowhead, demonstrates newly replicated episome. The hybridization probe was pRC/CMV.107/402-T. (E) To evaluate amplification of a cotransfected plasmid in concert with T antigen episomes, HT-1376 bladder carcinoma cells were transfected with T antigen expression vectors and a reporter replication plasmid containing the SV40 DNA origin, pSV2CAT. DNA harvested from cells 4 days after gene transfer was sequentially digested with BamHI to linearize pSV2CAT and then with DpnI. The hybridization probe was a BamHI–HindIII chloramphenicol acetyltransferase fragment. CMV, pRC/CMV transfectants (no T antigen); DC, DpnI digestion control consisting of 5 μg of genomic DNA and 2 ng of either pRC/CMV.107/402-T (D, lane 9) or pSV2CAT (E, lane 9).
107/402-T Does Not Bind to Wild-Type RB, p107, and p53 Proteins.
The biochemical correlate of SV40 large T antigen-mediated induction of tumorigenicity is complex formation with p53, RB, and possibly RB-related proteins, such as p107 (8–11). To directly evaluate the ability of 107/402-T to bind to wild-type RB, p107, and p53, in vitro translated wild-type and mutant T antigens were added to extracts from CV-1 cells in which human RB, p107, or p53 was transiently expressed at high levels. After addition of corresponding monoclonal antibodies recognizing these human tumor suppressor gene proteins, an immunoprecipitation analysis was performed (Fig. 1 B and C). Band intensities were scanned using a phosphorimager to quantitate binding interactions (Table 1). These data demonstrate that 107/402-T does not bind significantly to RB, p107, or p53.
Table 1.
Binding of wild-type and mutant SV40 large T antigens to RB, p107, and p53 tumor suppressor gene products
| Tumor suppressor gene product | T, % | Observed signal compared to T
|
||
|---|---|---|---|---|
| 107-T, % | 402-T, % | 107/402-T, % | ||
| RB | 100 | 0.03 | 67 | 0.07 |
| p107 | 100 | 0 | 79 | 0 |
| p53 | 100 | 36.2 | 0 | 0 |
107/402-T Is Replication-Competent.
The replication activities of wild-type and mutant SV40 large T antigens have been evaluated in a panel of human, simian, dog, hamster, rat, and murine cell lines (Table 2). An example of this analysis evaluating human hepatoma cell line Hep G2 and HT-1376 bladder carcinoma cells is shown in Fig. 1 D and E.
Significant replication activity was observed in human and simian lines, with less activity in a hamster cell line and no detectable activity in dog, rat, or murine samples. In Hep G2 cells, for example, a copy number of approximately 25,000 per cell was noted by 2 days after gene transfer, and copy numbers ranging from 80 to 100,000 were observed in other human cell types. In most cases, the replication activity of 107/402-T was at least as high as that observed for wild-type T antigen (data not shown). The lack of replication activity in rodent cell lines was not unexpected because these species are nonpermissive for SV40 infection and replication (21).
107/402-T Expression and Episome Copy Number.
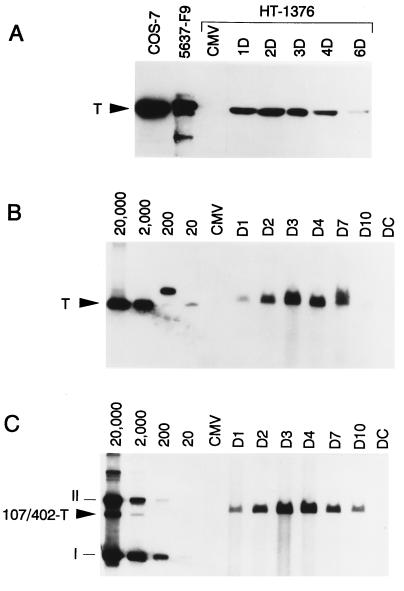
To design replicating vectors that yield superior expression characteristics, we evaluated the quantitative relationship between 107/402-T expression and the level of vector amplification. We transfected HT-1376 bladder cancer cells with pRC/CMV-T, pRC/CMV.107/402-T, or pRC/CMV, and cells were serially harvested for preparation of Western lysates or total cellular DNA.
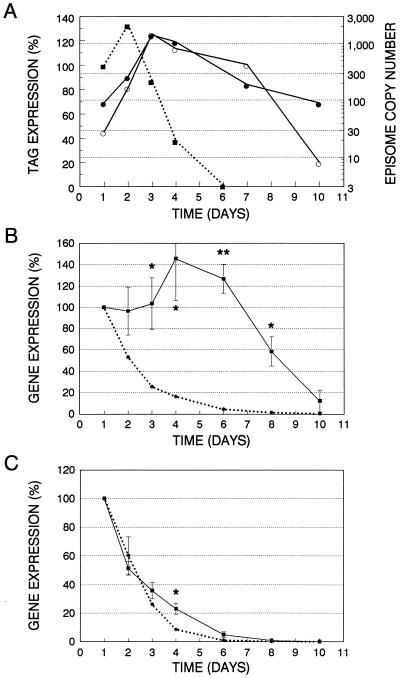
Presented in Fig. 2A is the time course of 107/402-T expression as a function of days after gene transfer. We observed an initial increase in gene expression followed by a decline by 3 days after transfection. Fig. 2 B and C shows Southern blots evaluating the copy number of wild-type and mutant T antigen episomes in HT-1376 transient transfectants. As predicted by the time course of 107/402-T expression, there is an initial exponential increase in episomal copy number followed by a gradual decline beginning on day 4 after gene transfer. The data in Fig. 2 are presented in graphic form in Fig. 3A. The exponential increase of vector copy number, indicated by the straight line on this semi-log plot, is due to the presence of significant levels of 107/402-T in these cells. However, as 107/402-T expression declines, the episomal plasmids stop replicating and the copy number per cell diminishes logarithmically in proportion to the doubling time of these cells (≈25 h). The biphasic shape of this curve suggests a threshold level of 107/402-T expression below which episomal replication ceases.
Figure 2.
107/402-T expression and episome copy number in transiently transfected HT-1376 cells. (A) At the indicated times (days 1–6), cell extracts were prepared for Western blot analysis of 107/402-T expression using anti-T monoclonal antibody 416. The 96-kDa 107/402-T protein is indicated by the arrowhead. Positive controls for T antigen expression include COS-7 and a clone of 5637 bladder carcinoma cells stably expressing 107-T (F9). (B and C) Southern blot analysis of HT-1376 cells transiently transfected with episomes encoding wild-type T or 107/402-T, respectively. At the indicated times (days 1–10), cells were harvested, and total cellular DNA (4 μg) was digested with both ApaI and DpnI. Linearized DNA (arrowhead) indicates the presence of replicated episome. A standard curve ranging from 20 to 20,000 copies per cell was calculated based on transfection efficiency and modal chromosome number. In B, standard curve plasmid DNA was linearized with ApaI, whereas uncut DNA was loaded in C. CMV, negative control pRC/CMV transfectants 3 days after gene transfer; DC, DpnI digestion control consisting of 4 μg of HT-1376 DNA and 2 ng of pRC/CMV.107/402-T.
Figure 3.
SV40-based episomal copy number and gene expression in transiently transfected HT-1376 cells. (A) Densitometric analysis of data shown in Fig. 2. 107/402-T expression (▪) is presented on a relative scale, with 100% indicating the observed level at day 1. Copy number of episomes encoding wild-type T (○) or 107/402-T (•) is presented on a log scale. (B and C) Plotted are the relative levels of β-galactosidase (▪) and luciferase (•) expression in cells cotransfected with pRSVlacZII, pCMVint-lux, and either pRC/CMV.107/402-T or pRC/CMV, respectively. Gene expression is normalized for the day 1 value, which is considered to be 100%. Shown are means ± SE of three separate experiments. Significance was determined using a paired, one-tailed Student’s t test. ∗, P < 0.05; ∗∗ P < 0.01.
SV40 107/402-T Episomes: Efficient Vertical Transfer in Replicating Tumor Cells.
To correlate episomal copy number with gene expression, a single 100-mm dish of HT-1376 cells was cotransfected with pRSVlacZII, pCMVint-lux, and a 5-fold molar excess of pRC/CMV.107/402-T. pRSVlacZII will replicate in cells expressing 107/402-T because it contains an SV40 DNA origin, whereas pCMVint-lux will be unable to replicate despite the presence of 107/402-T because it lacks an SV40 DNA origin. β-galactosidase and luciferase reporter gene expression is tabulated separately as relative light units per milligram of protein extract, and relative activities are compared with the day 1 level (Fig. 3B). As anticipated, we observed maintenance of β-galactosidase expression with time because amplification of pRSVlacZII will permit efficient vertical transfer of this plasmid during cell replication. In contrast, luciferase expression exponentially decreases with time because the copy number of pCMVint-lux falls by at least 50% with each cell division. In fact, the observed half-life of luciferase decay, 27 h, is essentially the same value as the doubling time of HT-1376 cells (25 h).
To confirm that the half-life of β-galactosidase activity in HT-1376 cells is not significantly prolonged compared with luciferase activity and to control for potential differences in promoter activity, HT-1376 cells were cotransfected with each reporter plasmid and pRC/CMV. As shown in Fig. 3C, both luciferase and β-galactosidase activities exponentially fall at the same rate, demonstrating that differences in reporter gene stability or promoter activity do not account for maintenance of β-galactosidase activity in the presence of 107/402-T.
SV40 107/402-T Episomes: High Level Gene Expression.
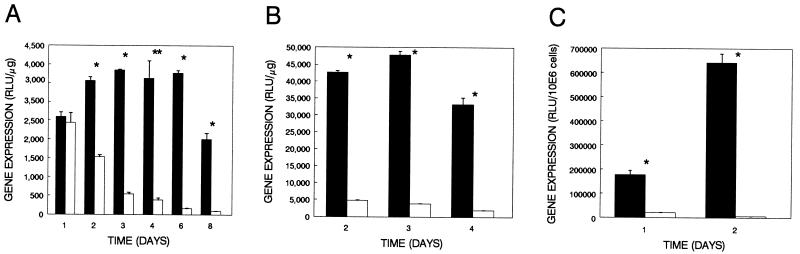
HT-1376 bladder, Hep G2 hepatoma, and RAJI lymphoma cells (Fig. 4) were transfected with pRSVlacZII and a 5-fold molar excess of either pRC/CMV.107/402-T or pRC/CMV, and the data are presented as absolute levels of β-galactosidase expression. There was an approximately 10-fold enhancement of episome-based gene expression by day 4 for HT-1376, a 20-fold enhancement by day 4 for Hep G2, and a 100-fold enhancement by day 2 for RAJI. These data demonstrate that our SV40-based expression system achieves significantly higher levels of gene expression than analogous, standard plasmid constructs.
Figure 4.
Episome-based gene expression in HT-1376 (A), Hep G2 (B), and RAJI (C). Cells were cotransfected with pRSVlacZII and either pRC/CMV.107/402-T (solid bars) or pRC/CMV (open bars). Shown are representative results from at least two separate experiments. Significance was determined using an unpaired, one-tailed Student’s t test. ∗, P < 0.0001; ∗∗, P = 0.0001.
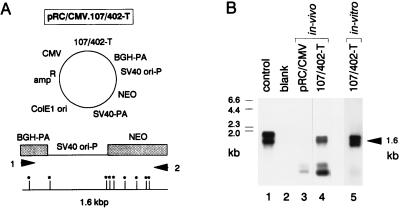
Gene Transfer In Vivo: Extrachromosomal Replication of 107/402-T Episomes.
To determine if our episomal vectors replicate extrachromosomally in vivo, we established conditions for efficiently transfecting established HT-1376 tumor xenografts by directly inoculating tumors with liposome/DNA complexes. To distinguish newly replicated episomal DNA from the larger amount of input DNA injected into the tumor nodule, we used a PCR-based episomal replication assay (20). The basis for this assay is illustrated in Fig. 5A for pRC/CMV.107/402-T. Explants were harvested for DNA preparation 3 days after gene transfer. Before PCR amplification, DNA samples were digested with DpnI. Because human cells lack DNA adenine methylase activity, DpnI will digest the input DNA used to transfect these cells but not newly synthesized DNA. Hence, only newly replicated, episomal DNA will be able to generate a PCR amplification product, as observed in the tumor explants transfected with pRC/CMV.107/402-T but not pRC/CMV (Fig. 5B). The redundancy of DpnI sites in the amplification fragment ensures that essentially all “input” DNA is inactivated before PCR amplification, as verified by the pRC/CMV-transfected tumor explant (Fig. 5B, lane 3). These data demonstrate that our SV40-based episomes replicate in human tumor cells after direct in vivo gene transfer.
Figure 5.
107/402-T-based episomes replicate extrachromosomally in human cells after direct in vivo gene transfer. (A) Map of pRC/CMV.107/402-T and the location of primers 1 and 2 used to amplify a 1.6-kbp region of this plasmid extending from the bovine growth hormone polyadenylylation site to the neomycin-resistance gene. Asterisks denote DpnI restriction sites. (B) PCR-based episomal replication assay. Southern blot analysis of PCR amplification products derived from HT-1376 cells transfected either in vivo (lanes 3 and 4) or in vitro (lane 5) with pRC/CMV or pRC/CMV.107/402-T. Before PCR amplification, 1 μg of total cellular DNA was digested with DpnI. Samples were then subjected to 32 cycles of PCR amplification. Amplification of 3 pg of pRC/CMV.107/402-T (positive control) is shown in lane 1, and the negative control, lacking template DNA, is shown in lane 2. The hybridization probe was generated from a pair of internally nested primers.
DISCUSSION
We have developed a novel, safety-modified episomal expression vector that replicates extrachromosomally in human cells and yields significantly higher levels of gene expression than standard, nonreplicating plasmid constructs. Our vectors are replication-competent in a wide spectrum of human tumors, as listed in Table 2. Because replication transactivators from DNA viruses possess significant transformation activity, construction of a replication-competent, transformation-negative SV40 T antigen mutant permits development of a new class of gene therapy vectors that yield very high levels of gene expression in unselected populations of transient transfectants. This vector system will, therefore, have particular utility for cancer gene therapy applications, in which high levels of transient expression of cytokines, immunogenic factors, and prodrug activators are predicted to yield enhanced effectiveness of tumor cell elimination.
The favorable expression characteristics of this vector system depends on reaching a threshold level of T antigen expression to drive extrachromosomal replication of the episomal plasmid. As illustrated in Fig. 2 for HT-1376 cells, only a short period of 107/402-T expression is needed to significantly amplify the episomal copy number because an increase of approximately 1 log in extrachromosomal copy number per day can be achieved. The time course of vector amplification likely depends on several factors, including the level and duration of 107/402-T expression and the availability of proteins that are required for cellular DNA replication. Host factors, including expression of c-myc or N-myc, may facilitate vector amplification (22–24). T antigen must also be phosphorylated to achieve replication competence (25). Other cell factors, such as p53 and RB, may inhibit vector replication by inactivating the replication-transactivator function of T antigen molecules that are bound to these host proteins (26–30). This latter effect may account, in part, for differences in the observed copy number of episomes encoding wild-type and mutant T antigen. For example, Hep G2 expresses wild-type p53 and RB (31), and, compared with pRC/CMV-T, these cells have approximately 100-fold higher levels of pRC/CMV.107/402-T, 80-fold higher levels of pRC/CMV.402-T, and 30-fold higher levels of pRC/CMV.107-T (Fig. 1D). Differences in the level of episomal copy number in Hep G2 may therefore reflect the relative contribution of p53 and RB in inhibiting T antigen function. In contrast, HT-1376 cells support approximately equal levels of vector amplification (Fig. 1E), consistent with the presence of both p53 and RB gene mutations in this cell line (17, 32). Hence, our data suggest that the copy number of episomes encoding 107/402-T will often be higher than episomes encoding wild-type T antigen because 107/402-T is not subject to down-modulation by either p53 or RB.
SV40 virus replicates in concert with the host cell (33), and episomes encoding wild-type T antigen likely amplify preferentially during S phase. This correlation may be regulated, in part, by cell cycle-associated phosphorylation of p53 and RB, and generation of replication-competent, unbound T antigen during S phase (34–37). In contrast, the replication activity of 107/402-T in transformed cells may be partially uncoupled from S phase because this molecule does not bind to either p53 or RB. This point is of some significance because the percentage of cells traversing S phase in human tumors varies considerably (38). Moreover, it is unclear if 107/402-T will have any replication activity in post-mitotic human cells because 107/402-T is unable to stimulate entry of simian cells into S phase (39). Hence, the replication properties of 107/402-T based episomes may be strongly linked to mitotically active cells.
The optimal number of transcriptional cassettes per cell to yield high levels of heterologous gene expression is currently unknown, although excessive copy numbers may limit gene expression due to induction of cellular toxicities by competition for replication enzymes, transcription factors, and possibly ribosomes. In addition, plasmids that are actively replicating may have diminished efficiencies of gene expression (40). Therefore, transient expression of 107/402-T may be optimal in amplifying the number of transcriptional cassettes to approximately 103 to 104 per cell, a level that appears from our data to yield high level gene expression without induction of cellular toxicities.
Transgenic mice will be useful to evaluate the potential transformation properties of 107/402-T. Tumor formation, if observed, might be mediated by rare back mutations in 107/402-T or inactivation of a putative cell protein targeted by an N-terminal T antigen domain (41). It is noteworthy, however, that transgenic mice expressing this N-terminal T antigen domain fail to develop tumors (42). Additionally, transgenic mice expressing the 5080 T antigen mutant (43), which is deficient in binding RB and p53 but is replication-incompetent, fail to develop cancers, whereas control mice expressing wild-type T antigen died from malignant brain tumors (44). Hence, these data suggest that 107/402-T has a low potential for inducing cell transformation. Moreover, the transformation potential of 107/402-T can be minimized further by using inducible promoter elements to regulate expression of 107/402-T. This modification in vector design will permit 107/402-T expression to be turned off 1–2 days after gene transfer, thereby permitting a transient boost in vector copy number. Tissue-specific promoter elements will also limit vector amplification and high level gene expression primarily to tumor cells. Finally, the predicted low risk of using 107/402-T based episomal vectors for cancer gene therapy will be justified in patients with terminal disease if these vectors increase effectiveness, as predicted from their expression characteristics.
Acknowledgments
We thank J. DeCaprio for plasmids pSG5-T and pSG5-K1; D. Simmons for SV40 mutant 402DE; P. Hobart for pCMVint-lux; L. Culp for pRSVlacZII; P. Davis for MDCK-2 cells; and R. Graham for BHK cells. M.J.C. was supported by American Cancer Society Grant IRG-186, Edison Biotechnology Center, Ohio Board of Regents, and National Institutes of Health Grant R55CA/OD66780. M.J.C. is a recipient of a Clinical Oncology Career Development Award from the American Cancer Society.
ABBREVIATIONS
- SV40
simian virus 40
- RB
retinoblastoma gene
- CMV
cytomegalovirus
- RSV
Rous sarcoma virus
References
- 1.Goff S P. Cancer Cells. 1990;2:172–178. [PubMed] [Google Scholar]
- 2.Biamonti G, Della Valle G, Talarico D, Cobianchi F, Riva S, Falaschi A. Nucleic Acids Res. 1985;13:5545–5561. doi: 10.1093/nar/13.15.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yates J L, Warren N, Sugden B. Nature (London) 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 4.Milanesi G, Barbanti-Brodano G, Negrini M, Lee D, Corallini A, Caputo A, Grossi M P, Ricciardi R P. Mol Cell Biol. 1984;4:1551–1560. doi: 10.1128/mcb.4.8.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper M J, Miron S M. Hum Gene Ther. 1993;4:557–566. doi: 10.1089/hum.1993.4.5-557. [DOI] [PubMed] [Google Scholar]
- 6.Tsui L C, Breitman M L, Siminovitch L, Buchwald M. Cell. 1982;30:499–508. doi: 10.1016/0092-8674(82)90247-1. [DOI] [PubMed] [Google Scholar]
- 7.Wilson J B, Levine A J. Curr Top Microbiol Immunol. 1992;182:375–384. doi: 10.1007/978-3-642-77633-5_48. [DOI] [PubMed] [Google Scholar]
- 8.Linzer D H, Levine A J. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 9.DeCaprio J A, Ludlow J W, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston D M. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 10.Ewen M E, Xing Y, Lawrence J B, Livingston D M. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 11.Claudio P P, Howard C M, Baldi A, De Luca A, Fu Y, Condorelli G, Sun Y, Colburn N, Calabretta B, Giordano A. Cancer Res. 1994;54:5556–5560. [PubMed] [Google Scholar]
- 12.Kalderon D, Smith A E. Virology. 1984;139:109–137. doi: 10.1016/0042-6822(84)90334-9. [DOI] [PubMed] [Google Scholar]
- 13.Lin J-Y, Simmons D T. J Virol. 1991;65:2066–2072. doi: 10.1128/jvi.65.4.2066-2072.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin W C, Culp L A. BioTechniques. 1991;11:344–351. [PubMed] [Google Scholar]
- 15.Manthorpe M, Cornefert-Jensen F, Hartikka J, Felgner P L, Rundell A, Margalith M, Dwarki V. Hum Gene Ther. 1993;4:419–431. doi: 10.1089/hum.1993.4.4-419. [DOI] [PubMed] [Google Scholar]
- 16.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Nature (London) 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 17.Cooper M J, Haluschak J J, Johnson D, Schwartz S, Morrison L J, Lippa M, Hatzivassiliou G, Tan J. Oncol Res. 1994;6:569–579. [PubMed] [Google Scholar]
- 18.Graham F L, Van der Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 19.Young D C, Kingsley S D, Ryan K A, Dutko F J. Anal Biochem. 1992;315:24–30. doi: 10.1006/abio.1993.1549. [DOI] [PubMed] [Google Scholar]
- 20.Cooper M J, Hatzivassiliou G, Miron S. BioTechniques. 1994;16:20–24. [PubMed] [Google Scholar]
- 21.Fried M, Prives C. Cancer Cells. 1986;4:1–16. [Google Scholar]
- 22.Classon M, Henriksson M, Sumegi J, Klein G, Hammaskjold M L. Nature (London) 1987;330:272–274. doi: 10.1038/330272a0. [DOI] [PubMed] [Google Scholar]
- 23.Hermeking H, Wolf D A, Kohlhuber F, Dickmanns A, Billaud M, Fanning E, Eick D. Proc Natl Acad Sci USA. 1994;91:10412–10416. doi: 10.1073/pnas.91.22.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee N G, Yamaguchi J, Subramanian K N. Oncogene. 1991;6:1161–1169. [PubMed] [Google Scholar]
- 25.McVey D, Brizuela L, Mohr I, Marshak D R, Gluzman Y, Beach D. Nature (London) 1989;341:503–507. doi: 10.1038/341503a0. [DOI] [PubMed] [Google Scholar]
- 26.Braithwaite A W, Horst-Werner S, Addison C, Palmer C, Rudge K, Jenkins J R. Nature (London) 1987;329:458–460. doi: 10.1038/329458a0. [DOI] [PubMed] [Google Scholar]
- 27.Gannon J V, Lane D P. Nature (London) 1987;329:456–458. doi: 10.1038/329456a0. [DOI] [PubMed] [Google Scholar]
- 28.Wang E H, Friedman P N, Prives C. Cell. 1989;57:379–392. doi: 10.1016/0092-8674(89)90913-6. [DOI] [PubMed] [Google Scholar]
- 29.Friedman P N, Kern S E, Vogelstein B, Prives C. Proc Natl Acad Sci USA. 1990;87:9275–9279. doi: 10.1073/pnas.87.23.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uzvolgyi E, Classon M, Henriksson M, Huang H-J S, Szekely L, Lee W-H, Klein G, Sumegi J. Cell Growth Differ. 1991;2:297–303. [PubMed] [Google Scholar]
- 31.Puisieux A, Galvin K, Troalen F, Bressac B, Marcais C, Galun E, Ponchel F, Yakicier C, Ji J, Ozturk M. FASEB J. 1993;7:1407–1413. doi: 10.1096/fasebj.7.14.8224613. [DOI] [PubMed] [Google Scholar]
- 32.Horowitz J M, Park S H, Bogenmann E, Cheng J C, Yandell D W, Kaye F J, Minna J D, Dryja T P, Weinberg R A. Proc Natl Acad Sci USA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePamphilis M L, Wassarman P M. In: Organization and Replication of Viral DNA. Kaplan A S, editor. Boca Raton, FL: CRC; 1982. pp. 37–114. [Google Scholar]
- 34.Bischoff J R, Friedman P M, Marshak D R, Prives C, Beach D. Proc Natl Acad Sci USA. 1990;87:4766–4770. doi: 10.1073/pnas.87.12.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milner J, Cook A, Mason J. EMBO J. 1990;9:2885–2889. doi: 10.1002/j.1460-2075.1990.tb07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sturzbecher H W, Maimets T, Chumakov P, Brain R, Addison C, Simanis V, Rudge K, Philp R, Grimaldi M, Court W, Jenkins J R. Oncogene. 1990;5:795–780. [PubMed] [Google Scholar]
- 37.Ludlow J W, Shon J, Pipas J M, Livingston D M, DeCaprio J A. Cell. 1990;60:387–396. doi: 10.1016/0092-8674(90)90590-b. [DOI] [PubMed] [Google Scholar]
- 38.Steel G G. Growth Kinetics of Tumors. Oxford: Clarenden; 1977. pp. 185–216. [Google Scholar]
- 39.Dickmanns A, Zeitvogel A, Simmersbach F, Weber R, Arthur A K, Dehde S, Wildeman A G, Fanning E. J Virol. 1994;68:5496–5508. doi: 10.1128/jvi.68.9.5496-5508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haase S B, Heinzel S S, Calos M P. Mol Cell Biol. 1994;14:2516–2524. doi: 10.1128/mcb.14.4.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaciuk P, Carter M C, Pipas J M, Moran E. Mol Cell Biol. 1991;11:2116–2124. doi: 10.1128/mcb.11.4.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saenz Robles M T, Symonds H, Chen J, Van Dyke T. Mol Cell Biol. 1994;14:2686–2698. doi: 10.1128/mcb.14.4.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peden K W C, Srinivasan A, Farber J M, Pipas J M. Virology. 1989;168:13–21. doi: 10.1016/0042-6822(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Tobin G J, Pipas J M, Van Dyke T. Oncogene. 1992;7:1167–1174. [PubMed] [Google Scholar]