Abstract
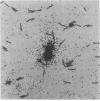
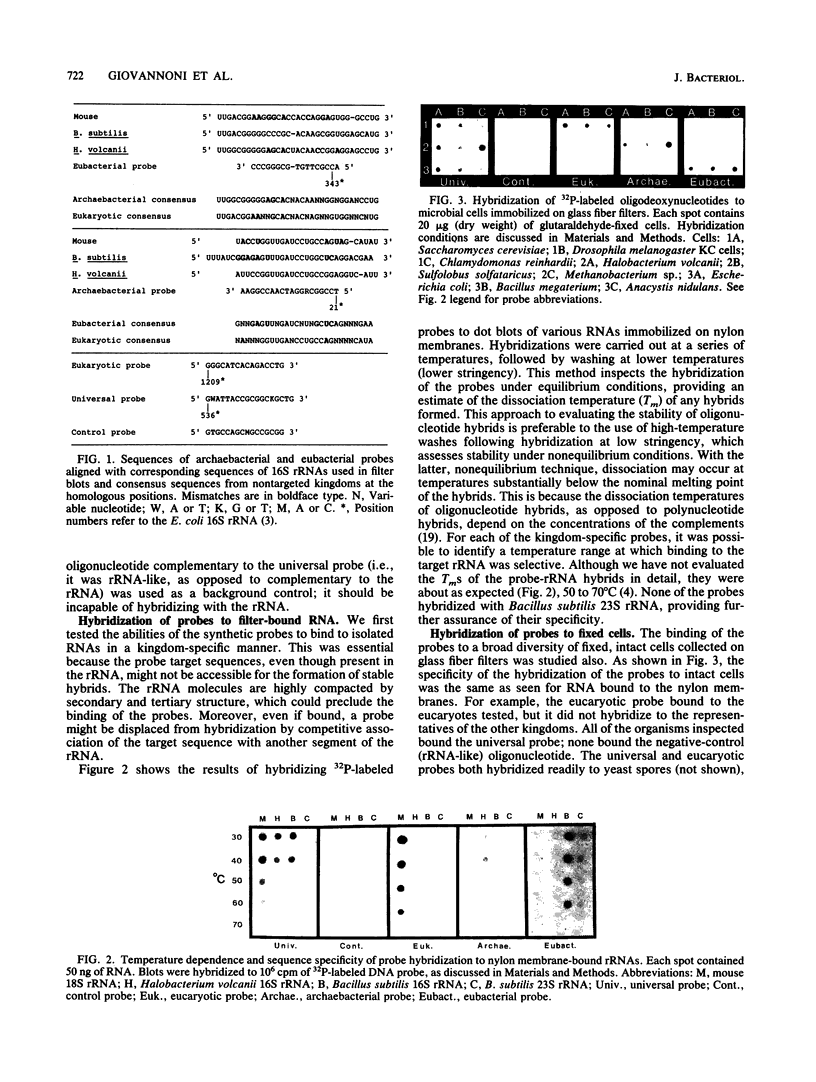
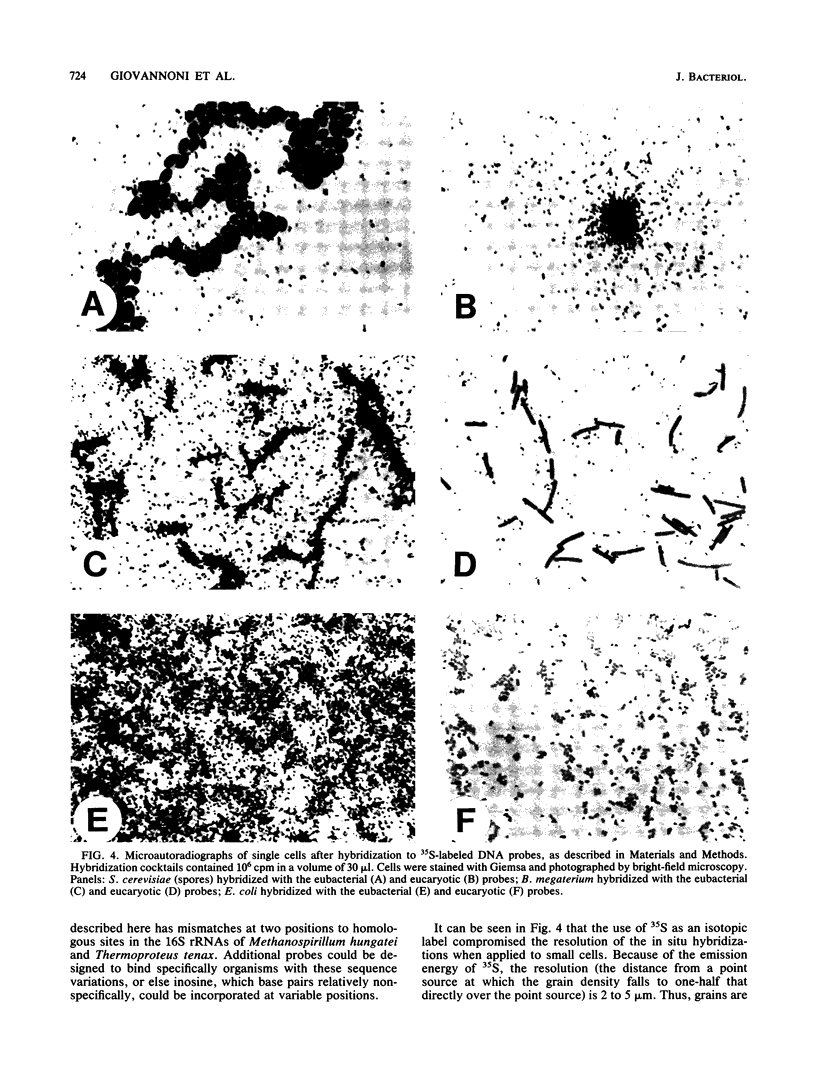
Examination of collections of 16S rRNA sequences revealed sequence domains that were unique to (and invariant within) the three primary lines of cellular descent: the archaebacteria, the eubacteria, and the eucaryotes. Oligodeoxynucleotides complementary to these conserved sequence domains were synthesized and used as hybridization probes. Each of the radiolabeled probes specifically hybridized to nylon membrane-bound 16S rRNA from the targeted kingdom. A probe complementary to a universally conserved sequence in 16S rRNAs was used as a positive control, while its complement provided a negative control for nonspecific binding. The abilities of the probes to bind specifically to whole, fixed cells representing a broad array of phylogenetic diversity were tested in whole-cell dot blot assays. Again, all of the probes specifically bound the targeted groups. By microautoradiography, the method was extended to permit phylogenetic identification of single cells microscopically.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brock T. D., Brock M. L. Autoradiography as a tool in microbial ecology. Nature. 1966 Feb 12;209(5024):734–736. doi: 10.1038/209734a0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Cohen L. W., Riggs A. D., Morin C., Itakura K., Richards J. H. Oligonucleotide-directed mutagenesis as a general and powerful method for studies of protein function. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6409–6413. doi: 10.1073/pnas.79.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echalier G., Ohanessian A. Isolement, en cultures in vitro, de lignées cellulaires diploïdes de Drosophila melanogaster. C R Acad Sci Hebd Seances Acad Sci D. 1969 Mar 31;268(13):1771–1773. [PubMed] [Google Scholar]
- Gorman D. S., Levine R. P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel U. B., Geiser A., Stanbridge E. J. Oligonucleotide probes complementary to variable regions of ribosomal RNA discriminate between Mycoplasma species. J Gen Microbiol. 1987 Jul;133(7):1969–1974. doi: 10.1099/00221287-133-7-1969. [DOI] [PubMed] [Google Scholar]
- Kuritza A. P., Salyers A. A. Use of a species-specific DNA hybridization probe for enumerating Bacteroides vulgatus in human feces. Appl Environ Microbiol. 1985 Oct;50(4):958–964. doi: 10.1128/aem.50.4.958-964.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Pace B., Matthews E. A., Johnson K. D., Cantor C. R., Pace N. R. Conserved 5S rRNA complement to tRNA is not required for protein synthesis. Proc Natl Acad Sci U S A. 1982 Jan;79(1):36–40. doi: 10.1073/pnas.79.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgaramella V., Khorana H. G. CXII. Total synthesis of the structural gene for an alanine transfer RNA from yeast. Enzymic joining of the chemically synthesized polydeoxynucleotides to form the DNA duplex representing nucleotide sequence 1 to 20. J Mol Biol. 1972 Dec 28;72(2):427–444. doi: 10.1016/0022-2836(72)90155-6. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]
- Yu S. M., Gorovsky M. A. In situ dot blots: quantitation of mRNA in intact cells. Nucleic Acids Res. 1986 Oct 10;14(19):7597–7615. doi: 10.1093/nar/14.19.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]