Abstract
CCAAT/enhancer binding protein (C/EBP) ɛ is a recently cloned member of the C/EBP family of transcription factors and is expressed exclusively in cells of hematopoietic origin. The human C/EBPɛ gene is transcribed by two alternative promoters, Pα and Pβ. A combination of differential splicing and alternative use of promoters generates four mRNA isoforms, of 2.6 kb and 1.3–1.5 kb in size. These transcripts can encode three proteins of calculated molecular mass 32.2 kDa, 27.8 kDa, and 14.3 kDa. Accordingly, Western blots with antibodies specific for the DNA-binding domain, that is common to all forms, identify multiple proteins. C/EBPɛ mRNA was greatly induced during in vitro granulocytic differentiation of human primary CD34+ cells. Retinoic acid treatment of HL60 promyelocytic leukemia cells for 24 hr induced C/EBPɛ mRNA levels by 4-fold, while prolonged treatment gradually reduced mRNA expression to pretreatment levels. Transient transfection experiments with expression vectors for two of the isoforms demonstrated that the 32.2-kDa protein is an activator of transcription of granulocyte colony-stimulating factor receptor promoter, while the 14.3-kDa protein is not. Thus, C/EBPɛ is regulated in a complex fashion and may play a role in the regulation of genes involved in myeloid differentiation.
Hematopoietic stem cells have the ability to promote continuous self-renewal by controlled proliferation and expansive differentiation into all cells of hematopoietic lineages. Regulation of the molecular mechanisms that direct cellular proliferation and differentiation during hematopoiesis depends, partly, on the action of certain transcription factors (1–3).
Several members of the basic region-leucine zipper (bZIP) class of transcription factors, including the CCAAT/enhancer binding protein (C/EBP) family have been implicated in the differentiation of hematopoietic cells (1–5). C/EBP proteins have highly conserved bZIP C-terminal regions, but differ in their N-terminal regions (6, 7). A consensus site, 5′-A4T3T2G1C0G0C1A2A3T4-3′, has been identified as the target for interactions between DNA and the C/EBP proteins that bind to such sites either as homodimers or heterodimers (8). Dimerization is a prerequisite for the ability of these proteins to bind DNA. To date, six mammalian members have been characterized, C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ, C/EBPɛ, and C/EBPζ (5, 6, 9, 10). C/EBPα, C/EBPβ, and C/EBPδ are activators of transcription and are able to transactivate a variety of target genes. In contrast, C/EBPγ and C/EBPζ lack transactivation domains and instead act as transdominant repressors of transcription (11, 12).
The high degree of similarity of the DNA-binding and dimerization domains of the C/EBP proteins allows for the formation of a variety of C/EBP homodimers or heterodimers. The kind of dimer is determined by the nuclear composition of the C/EBP proteins in a given cell and dictates the expression of a distinct set of genes. Thus, with a relatively small number of transcriptional regulators, the expression of a large number of target genes can be tightly controlled. Recent studies have suggested that interactions between C/EBP members and CREB/ATF (13), Fos-Jun family (14), and NF-κB proteins (15, 16) may occur, increasing the pool of available interacting transcription factors.
The human C/EBPɛ gene was recently cloned and its location assigned to chromosome 14 (9, 10). The murine C/EBPɛ gene is located on mouse chromosome 14 (17). To further understand the role of C/EBPɛ in the regulation of genes involved in hematopoiesis and to gain insight into the expression and function of C/EBPɛ, we have determined the structural organization of both the human and murine C/EBPɛ genes. Differential splicing and alternative use of promoters in the human locus generates a total of four C/EBPɛ transcripts of 2.6 kb and 1.3–1.5 kb in size. These transcripts encode three proteins of calculated molecular mass 32.2 kDa, 27.8 kDa, and 14.3 kDa. C/EBPɛ mRNA was greatly induced during in vitro granulocytic differentiation of human primary CD34+ cells. Retinoic acid treatment of HL60 and U937 cells for 24 hr induced C/EBPɛ mRNA levels by approximately 4-fold, while prolonged treatment gradually reduced mRNA expression to pretreatment levels. The granulocyte colony-stimulating factor (G-CSF) receptor gene is a critical gene for myeloid differentiation, particularly for the production of neutrophils (18). Transient transfection experiments with expression vectors for two C/EBPɛ protein isoforms and the myeloid-specific G-CSF receptor promoter reporter gene construct, suggest a role for C/EBPɛ in the regulation of G-CSF receptor, a critical gene for myelogenesis.
MATERIALS AND METHODS
Isolation of Human and Murine Genomic and cDNA Clones for C/EBPɛ.
A 400-bp XhoI/PstI and a 403-bp BamHI/XbaI 32P-labeled human cDNA fragment (9) were used to screen a 129/SVJ genomic mouse library (Stratagene) and several cDNA libraries (mouse thymus, Stratagene; human bone marrow, mouse spleen, mouse 7 day embryo, CLONTECH). Phage inserts were characterized by restriction mapping, sequence analysis, and Southern hybridization.
5′ Rapid Amplification of cDNA Ends (RACE)–PCR, Reverse Transcription (RT)–PCR, and Northern Blot Analysis.
The 5′ ends of both the Pα- and Pβ-initiated transcripts were identified using a 5′ RACE method in combination with the Marathon cDNA amplification kit (CLONTECH). The C/EBPɛ-specific oligonucleotide primers used in this study were as follows (see also Fig. 1): RY 44 (exon Iα specific) 5′- GGA AGG CGT GTC AAG GGA GG-3′; RY 46 (exon Iβ specific) 5′- GCT GGA AGA GAG AAG TGC TG-3′; RY 66 (exon IIα specific) 5′-ATG TGT GAG CAT GAG GCC TCC ATT-3′; RY 67 (exon Iβ specific) 5′-CCT CCC TTG ACA CGC CTT CC-3′; RY 68 (internal probe) 5′-TGA GAA CGC GCA GAG GCT GGC-3′; RY 72 (exon IIβ specific) 5′-TAG CTC CCT GGG CTG CTG TAG ATG CCA-3′; RY 74 (exon IIα specific) 5′-CCT TGA GCC TTC TGG CCT CAG GCG CTG-3′; RY 77 (exon III specific) 5′-CCC CCC ACG CCC TTG ATG AG-3′; RY 79 (exon IIβ specific) 5′-GGC TCC TCC TTC ACC ACC ACA GCC CTG-3′; and RY 83 (exon Iβ + IIβ specific) 5′-CAG CGC CTT CCT GTC TCC TCC CTT GAC ACG-3′.
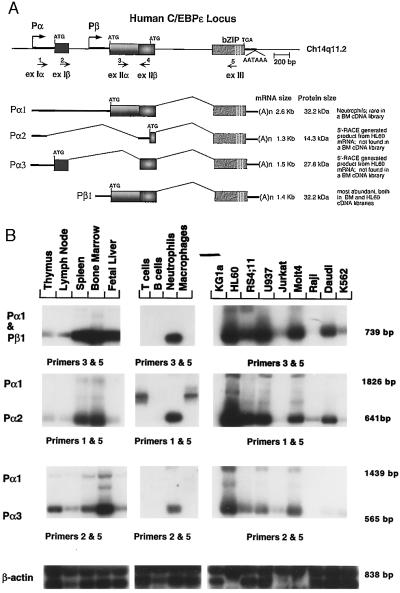
Figure 1.
Modular structure of the C/EBPɛ gene and its distinct RNA transcripts. (A) The transcription start sites of each of the two promoters, Pα and Pβ, are indicated by arrows. Boxes represent coding regions and thick lines represent untranslated regions of the mature mRNA. Nos. 1–5 correspond to primers RY46, RY44, RY66, RY68, and RY77, respectively. The location of the relevant start codons for each transcript and the common TGA stop codon is also shown. Vertical lines in the bZIP region indicate the presence of the leucine repeat. (B) RT-PCR reaction products amplified by the combination of C/EBPɛ specific primers indicated for each panel were analyzed by Southern blotting, using an internal primer as a probe (primer 4, A). All products were verified by sequence. β-Actin specific primers were used as controls. Fragment sizes are shown on the right, and the name of each transcript is at the left. The combination of primers 1 and 5 in primary T cells and macrophages amplifies a crossreacting product that is not related to epsilon based on sequence analysis of this fragment (Middle). A Pα1 RT-PCR-generated fragment of 1,826 bp is detected with longer exposure times in neutrophils (not shown).
We constructed cDNA libraries from HL60 (human promyelocytic leukemia) and MNFS 60 [myelogenous leukemia, monocyte CSF (M-CSF)-dependent, mouse] cell lines using 2 μg of poly(A)+ RNA as templates. The PCR primers used to detect C/EBPɛ mRNA expression were RY 66, RY 46, and RY 44 in combination with RY 77. Control β-actin-specific cDNA primers were from CLONTECH.
mRNA was extracted (19) from primary human T cells, B cells, neutrophils and macrophages, and several cell lines including human Molt-4 lymphoblastic leukemia, KG1a acute myelogenous leukemia, HL60, U937 monocytic leukemia, RS4;11 t(4;11) acute leukemia and Jurkat acute T cell leukemia. Human poly(A)+ RNA from bone marrow, fetal liver, lymph node, spleen, and thymus (CLONTECH) and cell lines Daudi, Raji Burkitt lymphomas, and K562 chronic myelogenous leukemia (CLONTECH) were also used as templates for first-strand synthesis.
Northern blots with approximately 20 μg of RNA per lane were hybridized (9), using two C/EBPɛ specific probes: a 3′ terminal 500-bp PstI fragment from a human cDNA clone (9) or a 306-bp PCR-generated fragment spanning nucleotides 3,254 to 3,560 (GenBank no. U48865U48865). To assure uniform loading, the blots were stripped and hybridized to oligonucleotides specific for either the 18S RNA, nucleotides 938–921 (20), or 28S RNA, nucleotides 4,036–4,020 (21).
Primary Cells, Cell Lines, and Transient Transfection Experiments.
Human primary CD34+ cells, peripheral blood monocytes, and neutrophils were isolated from healthy donors as described (22). CD34+-enriched cells were plated in human myeloid long-term culture medium supplemented with 300 units/ml G-CSF (G), 3 units/ml erythropoietin, or a combination of 1 ng/ml granulocyte-macrophage-CSF and 200 ng/ml M-CSF (GM + M) and cultured for 11 days (22). At the end of each differentiation experiment cells were stained by the Wright–Giemsa method, and differential cell counts were performed. U937, HL60, and K562 cells were grown in RPMI medium 1640 containing 10% fetal bovine serum and 2 nM l-glutamine. To induce in vitro erythrocytic differentiation, K562 cells were cultured in medium supplemented with 1.5% dimethyl sulfoxide (DMSO) for up to 5 days. U937 and HL60 cells were treated for 2 days with either 1.3 × 10−7 M phorbol 12-tetradecanoate 13-acetate (TPA) for monocytic differentiation or with 10−6 M retinoic acid for 4 days to induce granulocytic differentiation.
HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum and were transiently transfected at 70% confluency by liposome-mediated transfection reagent DOTAP (N-[1-(2,3)-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate) (Boehringer Mannheim). A DNA solution containing 5 μg of luciferase reporter plasmid pXP2 (23), (−74 to +67)G-CSFR-pXP2, Mut(−74 to +67)G-CSFR-pXP2 (24), and various amounts of C/EBP expression vectors (pCMV-C/EBPα, pCMV-C/EBPɛ32, pCMV-C/EBPɛ14) with 1 μg of β-galactosidase reporter plasmid (pCMV β) and pBluescript KS(+) plasmid DNA to adjust to a total of 10 μg of DNA per well was used. Luciferase activity was determined as previously described (25), and transfection efficiency was normalized to the levels of β-galactosidase expressed from 1 μg of cotransfected plasmid.
RESULTS
The C/EBPɛ Gene Uses Alternative Promoters and Differential Splicing to Generate Several RNA Isoforms.
Consistent with our previously described results (9), Northern blot hybridization using RNA from immune tissues and a C/EBPɛ-specific probe displayed a number of RNA isoforms of predominantly 1.3–1.5 and 2.6 kb in size (data not shown). To analyze each transcript isoform in detail, we screened a variety of cDNA and genomic libraries to isolate the human and the murine C/EBPɛ genes. We found that the C/EBPɛ gene was most abundant in bone marrow and HL60 cDNA libraries, although we have successfully isolated clones from mouse thymus and MNSF 60 libraries. Clones with different size inserts and unique or overlapping restriction enzyme patterns were isolated and sequenced. Several murine genomic C/EBPɛ clones were also isolated, covering almost 35.5 kb of the locus. We focused on the human locus and identified three exons by comparing genomic and cDNA clone sequences and by RT-PCR analysis and 5′ RACE–PCR using a variety of oligonucleotide combinations and poly(A)+ RNA templates. A schematic map of the human C/EBPɛ locus and all of the C/EBPɛ RNA transcripts are shown in Fig. 1A.
RNA was extracted from a variety of cells to identify the exon composition of each transcript by RT-PCR. To confirm that the PCR products were correctly amplified from a C/EBPɛ template, Southern blot experiments were performed using an internal C/EBPɛ specific primer as a probe. A representative set of results from such RT-PCR assays is shown in Fig. 1B, with the expected size shown on the right. mRNA from bone marrow, neutrophils, and HL60 cells amplified several C/EBPɛ fragments corresponding to C/EBPɛ isoforms, as did mRNA from spleen, fetal liver, U937, RS4;11, Molt4, and Daudi cells. A nonspecific RT-PCR-amplified product was detected in T cells and macrophages when primers 1 and 5 were used in combination (Fig. 1B). However, sequence analysis revealed that this fragment is not related to C/EBPɛ.
The presence of several differentially spliced C/EBPɛ mRNA species was confirmed by the isolation of 27 cDNA clones obtained from a human bone marrow cDNA library. The last exon encoding the bZIP domain was present in all cDNA clones; however, several differences were found in the 5′ end regions of the inserts. Pα3 contains exon 1, which is 436 bp long and includes two in-frame AUG initiation codons that are separated by 3 bp (see also Fig. 3B). Pα2 and Pα3 differ in that they use two different splice donor sites in exon 1 to splice to the same acceptor site in exon 2, as shown in Fig. 1A. Finally, the last exon of 334 bp, contains the bZIP domain of the molecule and is present in all C/EBPɛ isoforms.
Figure 3.
Three proteins with identical bZIP domains are encoded by the C/EBPɛ gene. (A) Western blot analysis of nuclear extracts from cell lines. Equivalent amounts of cell extracts from each of the cell lines shown were loaded. Molecular mass markers are shown on the left side. (B) The three proteins C/EBPɛ32, C/EBPɛ27, and C/EBPɛ14 are shown. Boxed residues represent identical amino acids. • indicate the position of each leucine residue of the zipper.
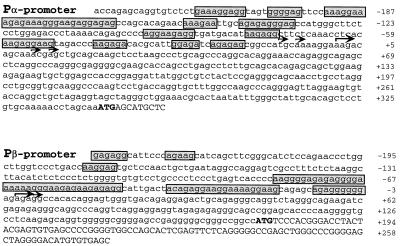
To determine the start site of the bone marrow cDNA clones we performed 5′ RACE assays. Sequence analysis of the bone marrow cDNA clones and 5′ RACE from mRNA derived from HL60 cells confirmed that they were genuine products of differentially spliced RNA transcripts. The start site identified in these clones was different due to the alternate use of two promoters, Pα and Pβ (Fig. 2). Thus, in addition to differential splicing, alternative use of promoters also contributes to the differences at the 5′ end region. Several 5′ RACE–PCR independent clones were sequenced to determine the initiation nucleotides shown for each promoter (Fig. 2, arrows). The equivalent promoter regions of human and murine genes were very conserved around the Pβ promoter but not the Pα promoter region (data not shown). Both Pα and Pβ promoter regions lack any obvious TATAAA or CAAT-like motifs. Similar to other TATA-less promoter regions of genes expressed in the hematopoietic system, a number of purine-rich stretches lie upstream of both start sites (Fig. 2). Several sites for the consensus sequences for PU.1 (an ets factor) RRRGAGGAAG were also found.
Figure 2.
Sequence of the two putative human C/EBPɛ promoter regions. The Pβ region is the major promoter used in HL60 cells, because the majority of the clones (>60%) identified by RACE–PCR were initiated at this site. The transcriptional start sites of the C/EBPɛ gene were determined by 5′-RACE–PCR and sequence analysis of several independent clones. Large arrows indicate sequence ends of the majority of cDNA clones for each promoter region. Small arrows show ends of less frequent RACE–PCR clones. The translation initiation codon is shown in uppercase and bold characters. Purine stretches are boxed.
Three Proteins Are Encoded by the C/EBPɛ Gene with Identical bZIP Domains.
C/EBPɛ protein was detected in cell lines of hematopoietic origin by Western blot analysis using a peptide antibody to the C-terminal region (Fig. 3A). Multiple size proteins were detected in MNFS 60, U937, and HL60. The four mRNA isoforms shown in Fig. 1A can give rise to three proteins that have the same basic DNA-binding domain but differ in the N-terminal part (Fig. 3). A band of varying intensity between experiments was also detected around 30 kDa. It may represent a degradation product of the larger protein or a nonspecific cross-reacting protein.
There are five in-frame AUG codons, but only three satisfy the Kozak context GCC(A/G)CCAUGG (26). Transcripts Pα1 and Pβ1 have identical reading frames with an ORF of 281 amino acids and have the capacity to generate two proteins of 32.2 kDa and 27.3 kDa. There is another in-frame AUG codon 32 amino residues downstream of the first (see Fig. 3B), However, initiation of translation at this downstream codon is less likely because of an imperfect match to the Kozak context (26). Furthermore, in vitro transcription–translation assays in rabbit reticulocyte lysates strongly favors initiation at the first AUG (data not shown).
Transcripts Pα2 and Pα3 can generate two proteins of 27.8 kDa and 14.3 kDa with the same basic domain as the previous proteins. Both AUG codons lie in a relatively strong Kozak context. Thus, several proteins can be encoded by the C/EBPɛ gene, and these proteins can be detected by Western blot analysis in MNFS60, U937, and HL60 (Fig. 3A). Two of the protein isoforms (32.3 kDa and 14.3 kDa) were used for transfection experiments (see below).
Induction of C/EBPɛ mRNA Expression Correlates with Granulocytic Differentiation.
Northern blot analysis of RNA extracted from human primary CD34+ cells showed that early progenitor cells do not express significant levels of C/EBPɛ mRNA (Fig. 4A, day 0). To test for steady-state C/EBPɛ mRNA levels during in vitro differentiation of primary human CD34+ cells enriched from peripheral blood of healthy donors, cells were plated in the presence of GM-CSF and M-CSF (GM + M), G-CSF (G) or erythropoietin (Epo). C/EBPɛ gene expression was detected in granulocytic cells after 8 days of culture and was greatly induced at day 11 of differentiation (Fig. 4A, G, 8 and 11). Control hybridizations of the same blot shows that the RNA samples in these two lanes have migrated evenly (28S control and data not shown). However, careful comparison between the two hybridizing bands at 8 and 11 indicated that the RNA form at day 11 is larger, possibly corresponding to a different C/EBPɛ isoform (see below). In accordance with the 5′-RACE results RNA extracted from HL60 cells displays multiple C/EBPɛ RNA isoforms (Fig. 4A; lane marked HL60). Finally, as shown in Fig. 4B, mature human neutrophils purified from peripheral blood of healthy donors express high levels of the long Pα1 isoform of C/EBPɛ whereas human monocytes do not express any detectable amounts of C/EBPɛ mRNA.
Figure 4.
(A) C/EBPɛ mRNA is preferentially expressed in granulocytic cells. Primary human CD34+ cells were enriched from peripheral blood of healthy donors and plated in medium supplemented with GM-CSF and M-CSF (GM + M; lanes 4–5), G-CSF (G; lanes 6–7), or erythropoietin (Epo, lanes 8–9). Northern blot was performed after 8 days (lanes 4, 6, 8) and 11 days (lanes 5, 7, 9) of culture. Lane 1, KG1a cells; lane 2, enriched CD34+ cells (83% CD34+); lane 3, primary CD34+ cells (CD34+) used for in vitro differentiation (lanes 4–9); lane 10, HL60 cells. (Upper) Hybridization to a C/EBPɛ-specific probe. (Lower) The blot was stripped and hybridized to a 28S RNA oligonucleotide control probe. (B) Mature human neutrophils express the long isoform of C/EBPɛ. Human monocytes and neutrophils were purified from peripheral blood of healthy donors. HeLa cells (lane 1), human monocytes (lane 2), human neutrophils (lane 3), and HL60 cells (lane 4) are shown. Migration of the 28S and 18S ribosomal RNA is shown to the left. (C) C/EBPɛ is up-regulated during granulocytic differentiation of human leukemic cell lines. K562 cells were cultured in medium supplemented with 1.5% DMSO (+DMSO) for 5 days to induce erythrocytic differentiation. U937 and HL60 cells were treated for 2 days with 1.3 × 10−7 M TPA (+TPA) for monocytic differentiation or with 10−6 M retinoic acid (+RA) for 4 days for granulocytic differentiation. Northern blot was performed at time points indicated above each lane, as described above with the exception that RNA was normalized to 18S ribosomal RNA (Lower).
We also tested whether C/EBPɛ is induced during granulocytic differentiation of several human leukemic cell lines. K562 cells were treated with 1.5% DMSO for 5 days to induce erythrocytic differentiation; U937 and HL60 cells were treated either with 1.3 × 10−7 M TPA to induce monocytic differentiation for 2 days, or with 10−6 M retinoic acid for 4 days for granulocytic differentiation. No expression of C/EBPɛ mRNA was detected in DMSO-treated or untreated K562 cells, consistent with the results obtained from the erythroid differentiation of primary CD34+ cells (Fig. 4C). In contrast, approximately a 4-fold induction of C/EBPɛ steady-state mRNA levels in HL60 cells was detected after retinoic acid treatment (Fig. 4C). However, this induction was transient because prolonged treatment with retinoic acid did not sustain these high levels (Fig. 4C, +RA 2–4). A similar induction was detected after 1-day treatment of U937 cells with retinoic acid (Fig. 4C). TPA treatment did not have any effect on the levels of C/EBPɛ RNA in U937 but it decreased the levels of C/EBPɛ RNA in HL60 cells. Thus, we conclude that C/EBPɛ is preferentially expressed during in vitro differentiation of primary cells to granulocytes, but not monocytes, and is strongly induced by retinoic acid in granulocytic cells.
The C/EBPɛ Gene Encodes Several Proteins with Different Transactivation Potential.
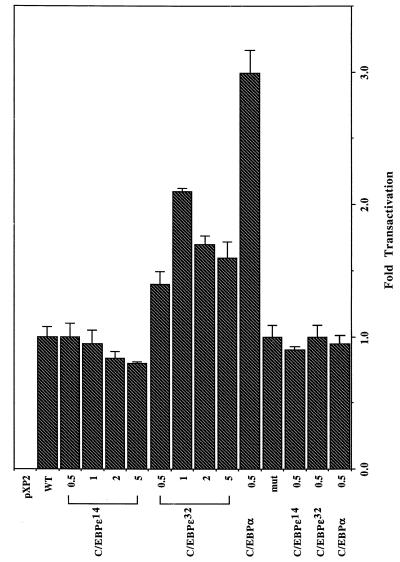
To test the transactivation potential of two of the C/EBP protein isoforms, the longest 32.2 kDa and the shortest 14.3 kDa, we generated pCMV-based expression constructs for both isoforms. The integrity of each expression construct was verified by sequence. These plasmids, a full-length C/EBPα expression construct and a wild-type and mutant G-CSF receptor-luciferase reporter, were used to transfect HeLa cells that do not express detectable levels of C/EBPα or C/EBPɛ. A cytomegalovirus-based lacZ expression vector was also used to normalize for transfection efficiency. As shown in Fig. 5, at low plasmid doses both C/EBPα and C/EBPɛ32 expression constructs were able to transactivate the G-CSF receptor promoter 3-fold and 2-fold, respectively. The transactivation of the G-CSF receptor promoter was relatively low, but this tranasctivation potential was consistently recorded over several independent experiments performed in duplicate and was not significantly affected by different plasmid DNA preparations. In contrast, C/EBPɛ14 failed to transactivate the reporter gene. Mutation of the C/EBP-binding site prevented activation by any of the C/EBP expression vectors. Thus, the short 14.3-kDa protein isoform lacks the transactivation potential of the longer variant.
Figure 5.
Activation of G-CSF receptor promoter-luciferase reporter construct by C/EBPα and C/EBPɛ in cotransfected HeLa cells. The G-CSF receptor-luciferase wild type (WT) and mutant (Mut) have been described (24). Increasing amounts of expression plasmids for C/EBPɛ or 0.5 μg of C/EBPα were cotransfected with a constant amount (5 μg) of both G-CSF receptor-luciferase reporter plasmids and 1 μg of β-galactosidase (pCMVβ) reporter. The amount of transfected DNA was adjusted with vector DNA to 10 μg per dish. The values are presented as a percentage of fold transactivation over the reporter. The experiments were repeated at least four times.
DISCUSSION
The C/EBP gene family includes six members characterized by highly conserved basic DNA binding and leucine zipper dimerization domains (7, 27, 28). In this study we describe several distinct features of the C/EBPɛ gene regulation and mRNA expression patterns that distinguish C/EBPɛ from the other C/EBP members. The C/EBPɛ gene is transcribed by two alternative promoters, Pα and Pβ, with a cell-type specificity. Differential splicing of the C/EBPɛ transcriptional unit generates several mRNA isoforms and adds to the complexity of the regulation of this gene. All isoforms share the same last exon that encodes for the bZIP carboxyl-terminal domain and have identical 3′ untranslated regions. However, the N-terminal regions of each protein are different. The structure of the mouse gene is similar to the human as are the sequences at the exon/intron boundaries and the Pβ promoter region (data not shown). Furthermore, there is a pronounced homology of 92.8% at the amino acid sequence level between the two 32.2-kDa protein isoforms (data not shown).
The deduced amino acid sequence of the longest human C/EBPɛ is 281 amino acids using Pα1 and Pβ1 RNA as translational templates. Alternative translational initiation from this RNA isoform is possible because there are three in-frame AUG codons (see Fig. 3B). Based on sequence comparisons between the different members of the C/EBP family we speculated earlier that C/EBPɛ is most related to C/EBPα (9). The homology starts in the N-terminal region of C/EBPα and the C/EBPɛ 32.2-kDa isoform, suggesting that the first start codon of this isoform is the dominant translational initiation codon. In vitro transcription/translation of a C/EBPɛ cDNA template encoding the Pα1 and Pβ1 form shows that the first upstream AUG codon is used efficiently in vitro, generating a protein of 32.3 kDa. Another protein is encoded by transcript Pα2, which is 129 amino acids long and has a calculated molecular mass of 14.3 kDa. Finally, a third protein that is 252 amino acids long and has a calculated molecular mass of 27.8 kDa is encoded by transcript Pα3. In several tissues and cell lines tested the Pβ1 transcript is by far the most abundant (this study and refs. 9 and 10). Thus, it is likely that the main molecular mechanism for control of C/EBPɛ gene expression is transcriptional regulation that favors initiation from the Pβ promoter region. This region is also the predominant promoter driving expression of the C/EBPɛ gene in mice.
Other C/EBP family members that lack introns use alternative translation initiation to generate functionally different factors. C/EBPβ or LAP can generate two proteins with distinct functions, LAP acts as an activator and LIP as a repressor that shares the same bZIP region with LAP but lacks the N-terminal activation domain identified in LAP (29). Similarly, C/EBPα is translated into multiple proteins with different transactivation potentials (30). All of these proteins share the same bZIP C-terminal domain but differ in their N-terminal amino acid sequences. A ribosome-scanning mechanism that selectively uses different in-frame AUGs to initiate translation from the same mRNA template has been suggested to explain the production of multiple C/EBPα proteins (30). Thus, N-terminally truncated forms are produced by several C/EBP family members, including C/EBPɛ, and are present in different vertebrate species such as rat, mouse, human, chicken, and Xenopus (30). Isoforms of transcriptional regulators that lack the transactivation domain are not uncommon among transcription factors that act as dimers. A heterodimer that contains an activator and a repressor will not be functional because only one activation domain is available. A variety of such examples have been described such as erbAα, mTFE3, Fos B, and I-POU (reviewed in ref. 31). Repressors can block transcriptional activation as homodimers by occupying an important regulatory element on a target gene. Several studies have demonstrated heterodimer formation between the C/EBP family members, including interactions of C/EBPɛ with C/EBPα, C/EBPβ, and C/EBPδ (27, 32, 33). However, the number of possible combinations of homodimers or heterodimers in a particular cell is reduced by the fact that the C/EBP genes are expressed in different cell types.
We reported previously that expression of the C/EBPɛ gene is restricted to tissues of the immune system (9). The present data demonstrate that expression of C/EBPɛ mRNA correlates with granulocytic differentiation of primary CD34+ cells. It is also detectable in CD34+CD33+ committed precursors and is highly expressed in CD11b+ individual myeloid bone marrow cells (D. Scadden, personal communication). Finally, C/EBPɛ mRNA is preferentially expressed in primary neutrophils. Retinoic acid treatment of HL60 promyelocytic cells to promote a granulocytic-like differentiation program greatly induces C/EBPɛ mRNA, suggesting a role for C/EBPɛ in this pathway. PU.1 and C/EBPα are both expressed early in myeloid differentiation (34). Both of these factors together with a nonidentified “band-X” factor have been shown to be involved in the regulation of the G-CSF receptor promoter (24). The transactivation potential of the long form of C/EBPɛ on G-CSF promoter reporter construct, although low under our experimental conditions, indicates that C/EBPɛ may also be involved in the regulation of this gene. The myeloperoxidase gene initiates transcription from two promoters separated by approximately 400 bp (35), and is a target of C/EBP proteins and PU.1 during granulocytic differentiation (36). C/EBPα, C/EBPβ, and C/EBPδ are expressed in an overlapping manner during in vitro granulocytic differentiation of A4 multipotential progenitor cells (36). Based on its expression pattern C/EBPɛ may also interact with the myeloperoxidase promoter regions.
The functional versatility of the C/EBPɛ gene is mirrored by the number of C/EBPɛ RNA isoforms that are produced in a cell- and tissue-specific manner. Alternative use of promoters and differential splicing of the primary transcriptional unit, unique among the C/EBP family members, results in ORFs that change the characteristics of the protein and may modulate its physiological role. Such molecular mechanisms are shared by other transcriptional regulators with modular structures such as CREM (37), CREB (38), and HLF (39). The modulatory structure of the C/EBPɛ gene may also be an indication of its evolutionary origin, perhaps reflecting an assembly of subunits with distinct function. The use of alternative promoter regions that are somewhat related may indicate that a duplication of the first modular unit has recently occurred. It is a tantalizing possibility that C/EBPɛ may be the evolutionary remnant of a primordial C/EBP gene with a modular structure.
The significance of the C/EBPɛ gene as an important regulator of the differentiation process in hematopoiesis has not yet been demonstrated in vivo. Our data suggest that this protein may be important in the regulation of certain genes such as the G-CSF receptor and plays a role in granulocytic differentiation. The ultimate demonstration of the role of C/EBPɛ in the hematopoietic system may have to wait for a functional analysis of this gene using the powerful tools of mouse genetics.
Acknowledgments
We are grateful to Drs. C. Barlow, D. Nelson, and A. Wynshaw-Boris for critical review of the manuscript and to Dr. R. Michael Blaese for generous support. We also thank Dr. Jeffrey Marx for help with CD34+ cell isolation. D.G.T. was supported by National Institutes of Health Grants CA41456 and DK48660.
Footnotes
Abbreviations: C/EBP, CCAAT/enhancer binding protein; G-CSF, granulocyte colony-stimulating factor; M-CSF, monocyte colony-stimulating factor; bZIP, basic region-leucine zipper; RT, reverse transcription; RACE, rapid amplification of cDNA ends; DMSO, dimethyl sulfoxide; TPA, phorbol 12-tetradecanoate 13-acetate.
References
- 1.Ness S A, Engel J D. Curr Opin Genet Dev. 1994;4:718–724. doi: 10.1016/0959-437x(94)90139-t. [DOI] [PubMed] [Google Scholar]
- 2.Shivdasani R A, Orkin S H. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 3.Tenen, D. G., Hromas, R., Licht, J. & Zhang, D.-E. (1997) Blood, in press. [PubMed]
- 4.Scott L M, Civin C I, Rorth P, Friedman A D. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 5.Wedel A, Ziegler-Heitbrock H W L. Immunobiology. 1995;193:171–185. doi: 10.1016/s0171-2985(11)80541-3. [DOI] [PubMed] [Google Scholar]
- 6.Johnson P F, McKnight S L. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 7.Xanthopoulos K G, Mirkovitch J. Eur J Biochem. 1993;216:353–360. doi: 10.1111/j.1432-1033.1993.tb18152.x. [DOI] [PubMed] [Google Scholar]
- 8.Koldin B, Suckow M, Seydel A, Wilken-Bergmann B V, Muller-Hill B. Nucleic Acids Res. 1995;23:4162–4169. doi: 10.1093/nar/23.20.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antonson P, Stellan B, Yamanaka R, Xanthopoulos K G. Genomics. 1996;35:30–38. doi: 10.1006/geno.1996.0319. [DOI] [PubMed] [Google Scholar]
- 10.Chumakov A M, Grillier I, Chumakova E, Chih D, Slater J, Koeffler H P. Mol Cell Biol. 1997;17:1375–1386. doi: 10.1128/mcb.17.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper C, Henderson A, Artandi S, Avitahl N, Calame K. Nucleic Acids Res. 1995;23:4371–4377. doi: 10.1093/nar/23.21.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barone M V, Crozat A, Tabaee A, Philipson L, Ron D. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- 13.Vinson C R, Hai T, Boyd S M. Genes Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 14.Hsu W, Kerpolla T K, Chen P L, Curran T, Chen-Kiang S. Mol Cell Biol. 1994;14:268–276. doi: 10.1128/mcb.14.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeClair K P, Blanar M A, Sharp P A. Proc Natl Acad Sci USA. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein B, Cogswell P C, Baldwin A S J. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins N C, Gilbert D J, Cho B C, Strobel M C, Williams S C, Copeland N G, Johnson P F. Genomics. 1995;28:333–336. doi: 10.1006/geno.1995.1150. [DOI] [PubMed] [Google Scholar]
- 18.Lui F, Wu H Y, Wesselsschmidt R L, Kornaga T, Link D C. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Torczynski R M, Fuke M, Bollon A P. DNA. 1985;4:283–291. doi: 10.1089/dna.1985.4.283. [DOI] [PubMed] [Google Scholar]
- 21.Barbu V, Dautry F. Nucleic Acids Res. 1989;17:7115. doi: 10.1093/nar/17.17.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H-M, Zhang P, Voso M T, Hohaus S, Gonzalez D A, Glass C K, Zhang D-E, Tenen D G. Blood. 1995;85:2918–2928. [PubMed] [Google Scholar]
- 23.Nordeen S K. BioTechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 24.Smith L T, Hohaus S, Gonzalez D A, Dziennis S E, Tenen D G. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 25.Antonson P, Pray M G, Jacobsson A, Xanthopoulos K G. Eur J Biochem. 1995;232:397–403. doi: 10.1111/j.1432-1033.1995.397zz.x. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Z, Umek R M, McKnight S L. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 28.McKnight S L. In: Transcriptional Regulation. McKnight S L, Yamamoto K, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 771–795. [Google Scholar]
- 29.Descombes P, Schibler U. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 30.Ossipow V, Descombes P, Schibler U. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foulkes N S, Sassone-Corsi P. Cell. 1992;68:411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- 32.Poli V, Mancini F P, Cortese R. Cell. 1990;63:643–653. doi: 10.1016/0092-8674(90)90459-r. [DOI] [PubMed] [Google Scholar]
- 33.Williams S C, Cantwell C A, Johnson P F. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 34.Voso M T, Burn T C, Wulf G, Lim B, Leone G, Tenen D G. Proc Natl Acad Sci USA. 1994;17:7932–7936. doi: 10.1073/pnas.91.17.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman A D, Krieder B L, Venturelli D, Rovera G. Blood. 1991;78:2426–2432. [PubMed] [Google Scholar]
- 36.Ford A, Bennett C A, Healy L Y, Towatari M, Greaves M F, Enver T. Proc Natl Acad Sci USA. 1996;93:10838–10843. doi: 10.1073/pnas.93.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laoide B M, Foulkes N S, Schlotter F, Sassone-Corsi P. EMBO J. 1993;12:1179–1191. doi: 10.1002/j.1460-2075.1993.tb05759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruppert S, Cole T J, Boshart M, Schmid E, Schutz G. EMBO J. 1992;4:1503–1512. doi: 10.1002/j.1460-2075.1992.tb05195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falvey E, Fleury-Olella F, Schibler U. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]