Abstract
The crisis in antibiotic resistance has resulted in an increasing fear of the emergence of untreatable organisms. Resistance to the glycopeptide antibiotic vancomycin in the enterococci, and the spread of these pathogens throughout the environment, has shown that this scenario is a matter of fact rather than fiction. The basis for vancomycin resistance is the manufacture of the depsipeptide d-Ala-d-lactate, which is incorporated into the peptidoglycan cell wall in place of the vancomycin target d-Ala-d-Ala. Pivotal to the resistance mechanism is the production of a d-Ala-d-Ala ligase capable of ester formation. Two highly efficient depsipeptide ligases have been cloned from vancomycin-resistant enterococci: VanA and VanB. These ligases show high amino acid sequence similarity to each other (≈75%), but less so to other d-Ala-d-X ligases (<30%). We have cloned ddls from two glycopeptide-producing organisms, the vancomycin producer Amycolatopsis orientalis and the A47934 producer Streptomyces toyocaensis. These ligases show strong predicted amino acid homology to VanA and VanB (>60%) but not to other d-Ala-d-X ligases (<35%). The d-Ala-d-Ala ligase from S. toyocaensis shows d-Ala-d-lactate synthase activity in cell-free extracts of S. lividans transformed with the ddl gene and confirms the predicted enzymatic activity. These results imply a close evolutionary relationship between resistance mechanisms in the clinics and in drug-producing bacteria.
Enterococci are frequent causes of nosocomial infections and are intrinsically resistant to many antibiotics, being susceptible only to synergistic penicillin/aminoglycoside therapy or to the glycopeptide antibiotic vancomycin (1). The recent emergence of resistance to vancomycin in these organisms (2, 3) has been greeted with grave alarm, not only because of the immediate problem of how to treat an infection when no other good drugs are available, but also because vancomycin-resistant enterococci (VRE) may serve as a reservoir for VanR genes, which will no doubt eventually make their way into more virulent organisms such as those of the genus Staphylococcus. High-level resistance to vancomycin in VRE (minimal inhibitory concentration > 1,000 μg/ml) is conferred by five genes located on a transposable element (4). These genes include vanR and vanS, which encode a two-component regulatory system that directs the transcription of three structural genes: vanA, vanH, and vanX. VanH is a d-specific α-ketoacid dehydrogenase that makes d-lactate (d-Lac) (5), VanA is a d-Ala-d-Ala ligase (Ddl) that catalyzes the synthesis of d-Ala-d-Lac (5), and VanX is a dipeptidase that clears cytoplasmic d-Ala-d-Ala synthesized by the endogenous Ddl (6, 7). Vancomycin acts by binding to the terminal d-Ala-d-Ala moiety of the bacterial peptidoglycan thus interfering with normal cell wall growth. As a consequence of the incorporation of d-Ala-d-Lac by VRE into the peptidoglycan layer in place of d-Ala-d-Ala, the affinity of vancomycin for the peptidoglycan diminishes over 1,000-fold, resulting in clinical antibiotic resistance (5).
The origin of the VanR genes has been the subject of some speculation. Related to this question is the increasing debate concerning the prudence of the continued agricultural use of glycopeptide antibiotics such as avoparcin (8–12) in view of the capacity for selection of VRE in the environment (13–15). That environmental VRE isolates have not been observed yet in North America where glycopeptides are not used in agriculture (16), unlike the situation in Europe, adds another dimension to the debate.
The amino acid sequence of the vancomycin-resistance enzyme VanA (17) and the related VanB (18) shows between 20% and 35% sequence homology to other Ddls, including the d-Ala-d-Lac ligases from organisms that are intrinsically resistant to vancomycin such as Leuconostoc spp. and Lactobacillus spp. (19). As part of an ongoing study of the biosynthesis of glycopeptide antibiotics, we have cloned the complete ddl gene from the glycopeptide-producing organism Streptomyces toyocaensis NRRL 15009, which produces the aglycopeptide A47934, and a partial ddl gene from Amycolatopsis orientalis, which produces vancomycin (Fig. 1). These genes encode proteins that show a high degree of homology to the VanA and VanB ligases and suggest that clinical vancomycin resistance may have originated in glycopeptide-producing organisms.
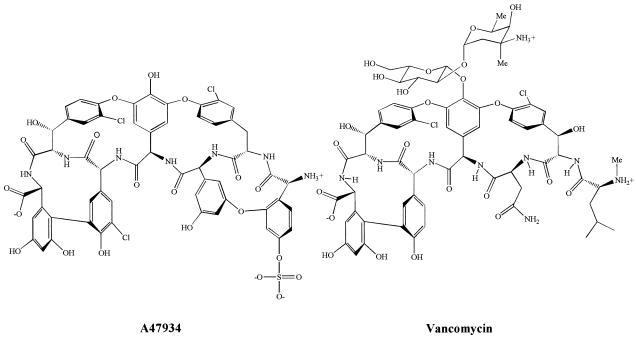
Figure 1.
Structures of glycopeptide antibiotics A47934 and vancomycin.
MATERIALS AND METHODS
Cloning of ddl Genes.
Cloning of ligase genes was achieved by PCR amplification using degenerate deoxyoligonucleotide primers designed to amplify an internal ddl fragment of approximately 650 bp (20). The primers were adjusted to account for the G/C preference of actinomycetes: 5′-GGIGAGGACGGIT(A)C(G)IC(A)TICAGGG-3′, 5′-GTGAAICCG(C)GGIAT(G)IGTGTT-3′. Each polymerase reaction contained genomic DNA as template (500 ng), Taq polymerase, 0.4 mM dNTPs, 1 μM primers, and 5.0% dimethyl sulfoxide. The putative 650-bp ddl fragments amplified from S. toyocaensis NRRL 15009 and A. orientalis C329.2 genomic DNA were separately cloned into pGEM-T (Promega) and sequenced in both directions by cycle sequencing methods on an Applied Biosystems automated DNA sequencer. For the A. orientalis internal 650-bp ddl fragment, further cloning and sequence analysis of the entire gene was not pursued. To isolate a clone encompassing the complete S. toyocaensis ddl gene as well as upstream and downstream flanking DNA, the 650-bp PCR fragment was randomly labeled with 32P and served as probe for Southern hybridization of genomic DNA isolated from S. toyocaensis, which was digested to completion using several restriction enzymes in separate experiments. Hybridizing fragments of DNA from digestion with SacI and BamHI were cloned into pBluescript II KS+ (Stratagene) and pGEM-7Zf (Promega), respectively, and confirmed to contain the ddl gene by colony hybridization. A 3.6-kb BamHI fragment was completely sequenced using a series of overlapping nested deletions created by ExoIII/S1 nuclease treatment. However, because of an internal BamHI site in the ddlM gene, sequence analysis from the overlapping 3-kb SacI fragment was required to complete the gene sequence.
d-Ala-d-X Activity Assays.
The 3-kb SacI fragment was subcloned into plasmid pFD666 (21) to give pFD666-Stoyddl, which was used to transform Streptomyces lividans 1326 protoplasts. Cultures (100 ml) of S. lividans 1326 bearing plasmid pFD666 (negative control) or pFD666-Stoyddl were grown for 48 hr in streptomycete antibiotic activity medium (per liter: soytone 15 g, d-glucose 15 g, NaCl 5 g, yeast extract 1 g, CaCO3 1 g, glycerol 2.5 ml) supplemented with neomycin (1 μg/ml), harvested, and lysed by two passages through a French pressure cell. The cell debris was removed by centrifugation and the supernatant (S1) recovered. d-Ala-d-Ala formation assay contained ATP (1 mM), d-Ala (10 mM), 40 mM KCl, d-[14C]Ala (2.2 × 105 dpm), 50 mM Tris⋅HCl at pH 8.6, and 150 μg of S1 protein. d-Ala-d-Lac formation assay contained ATP (1 mM), d-Ala (10 mM), 40 mM KCl, d-[14C]Lac (2.2 × 105 dpm), 50 mM Tris⋅HCl at pH 8.6, and 150 μg of S1 protein. Mixtures were incubated overnight at ambient temperature, applied to a cellulose TLC plate, and developed as previously described (5).
RESULTS AND DISCUSSION
Cloning of ddl Genes from Glycopeptide Producers.
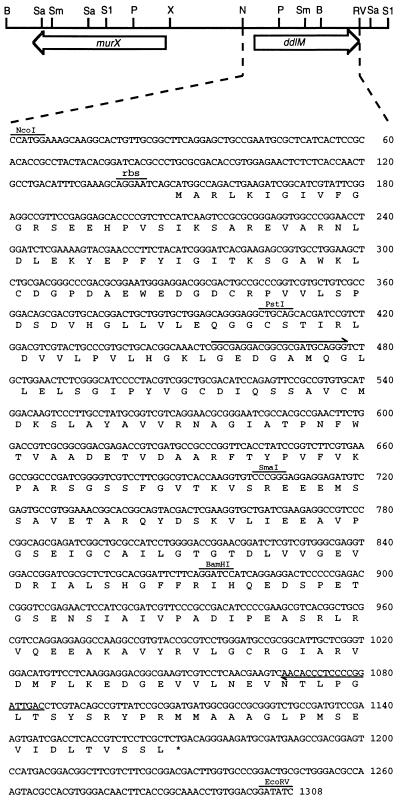
We used a PCR strategy based on the use of degenerate DNA primers that encode regions of homology between all Ddls, an approach that has proven successful for the cloning of other Ddls (e.g., ref. 20). DNA fragments of ≈650 bp were amplified from both S. toyocaensis and A. orientalis genomic DNA. Sequencing of these amplified products revealed strong amino acid homology to Ddls. We therefore cloned the entire gene from S. toyocaensis by standard methods. Complete sequencing of a 4.2-kb region of the genome revealed two ORFs (Fig. 2). The first ORF, murX, was 1,330 bases in length. Translation and blast (22) search revealed strong similarity to the Escherichia coli MurF protein, the UDP-N-acetylmuramoylalanyl-d-glutamyl-2,6-diaminopimelate d-alanine-d-alanine ligase [d-Ala-d-Ala adding enzyme, P(N) = 3.3 × 10−46]. The second ORF, ddlM, was 1,023 base pairs in length and blast search revealed high homology to d-Ala-d-Ala ligases [P(N) = 10−15 to −155].
Figure 2.
Restriction endonuclease map of the region cloned from S. toyocaensis containing ddlM and murX and the complete sequence of the ddlM gene, arrows indicate sites complementary to the PCR primers. Sa, SalI; Sm, SmaI; S1, SstI; P, PstI; N, NcoI; B, BamHI; RV, EcoRV; X, XmnI.
Sequence Analysis of ddl Gene Products.
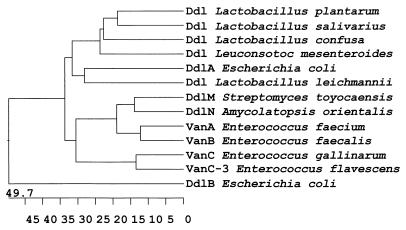
The S. toyocaensis ddl gene was found to encode a 341 amino acid protein, DdlM, with a predicted molecular mass of 36.7 kDa (Fig. 2). Alignment of the DdlM amino acid sequence with other Ddl proteins revealed notable homology (20–35% similarity) to the peptide synthesizing d-Ala-d-Ala ligases DdlA, DdlB, and VanC (Fig. 2a). The homology was, however, especially significant with the d-Ala-d-Lac ligases VanA and VanB from VRE (>60% similarity) but was moderate (≈25%) with the depsipeptide-synthesizing ligases from intrinsically vancomycin-resistant Gram-positive bacteria such as Leuconostoc mesenteroides and the Lactobacilli. A 600-bp segment of the ddl gene from the vancomycin producer A. orientalis was also cloned and sequenced (Fig. 3). The translated amino acid sequence, DdlN, was highly similar to the DdlM (72% similarity) and the VanA and VanB enzymes (60% and 62% respective similarity), thus these four enzymes form a distinct subfamily of Ddls (Fig. 4).
Figure 3.
Partial sequence of the ddl gene from A. orientalis C329.2.
Figure 4.
Phylogenetic relationship among Ddls. Phylogenetic tree prepared using the Clustal V method (23), the gap penalty and gap length were set to 10. The point accepted mutations (PAM250) weight table (24) was used to define the similarity between sequences (number of matching residues divided by the sum of the mismatched residues + matching residues + gapped residues) using the MegAlign software by DNAstar (Madison, WI). The horizontal axis is the percentage distance between sequences.
The crystal structure of the E. coli ligase, DdlB, complexed with ADP and a phosphorylated phosphinate inhibitor (25), along with recent mutagenesis experiments (26, 27), have revealed several important structural conditions required for DdlB catalysis. A flexible ω-loop closes off the active site upon substrate binding and brings two critical residues, Lys-215 and Tyr-216, into the active site. Lys-215 is conserved in all chromosomally encoded ligases, including VanC and the depsipeptide synthases from Leuconostoc and Lactobacillus (though not L. plantarum where this residue is a Met). Lys-215 binds the γ-phosphate of ATP and assists in phosphoryl transfer to form the d-Ala-1 acylphosphate intermediate (25, 26). Tyr-216 also plays a critical role in d-Ala-d-Ala synthesis. Closing of the ω-loop results in establishment of a hydrogen bond network from the phenolic hydroxyl of Tyr-216 through Ser-150 to Glu-15, which correctly positions d-Ala-1 for catalysis by interaction of Glu-15 with the α-amino group. Tyr-216 is replaced by a Phe residue in d-Ala-d-Lac ligases from Lactobacillus and Leuconostoc, and recently a Tyr-216 → Phe mutant in DdlB has been shown to be capable of d-Ala-d-Lac synthesis (27).
The precise positioning of the ω-loop in the acquired d-Ala-d-Lac ligases VanA and VanB is difficult to ascertain as the primary sequence homology breaks down in this region (Fig. 5a). However, biochemical evidence for the presence of such a loop in VanA has been established (28). These amino acid sequence differences in the ω-loop region highlight the unique nature of VanA and VanB and place these enzymes in a separate class that is distinct from other known Ddls. The fact that DdlM and DdlN share dramatic sequence homology with VanA and VanB in the ω-loop region clearly demonstrates a close evolutionary relationship between the ligases acquired by enterococci and those found in glycopeptide-producing organisms. The Ddls from producing organisms, like VanA and VanB, share broad sequence homology outside the ω-loop region, especially in substrate binding regions (Fig. 5 b and c). Thus the ω-loop structure is a distinguishing feature common to the Ddls from producing organisms, VanA, and VanB.
Figure 5.
Sequence overlaps in selected regions of Ddls. (a) Sequence overlap in the ω-loop region. (b) Sequence overlap at the N-terminal d-Ala1 binding region. (c) Sequence overlap in the Mg2+ binding region.
DdlM Ddl Activity.
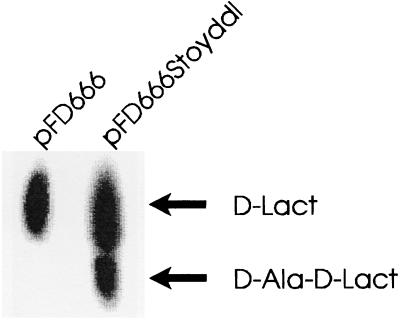
The close similarity at the amino acid level between Ddls from glycopeptide-producing organisms and the vancomycin-resistance enzymes of enterococci implied that they also shared similar substrate specificities. Cloning of ddlM into glycopeptide-sensitive S. lividans 1326 permitted assay of ligase activity in cell-free extracts. Like VanA, DdlM is a depsipeptide synthase capable of d-Ala-d-Lac biosynthesis (Fig. 6) as assessed by comigration with authentic VanA derived d-Ala-d-Lac (not shown). However, the presence in S. lividans of the ddlM gene did not confer resistance to A47934 or vancomycin at 0.5 μg/ml. This is probably due to a requirement for a VanX analogue to cleave endogenous S. lividans d-Ala-d-Ala as is the case for enterococci (29).
Figure 6.
d-Ala-d-X activity assays. The ddlM gene was cloned into plasmid pFD666 to give pFD666-Stoyddl and used to transform glycopeptide-sensitive S. lividans 1326. Assays were performed as previously described (5), and the products separated on cellulose TLC with 1-butanol:acetic acid:water (12:3:5) as solvent. d-Ala-d-Lac was identified by comigration with authentic depsipeptide produced by VanA.
Implications of This Study.
DdlM and DdlN are highly similar in amino acid sequence and enzymatic specificity to the recently emergent enzymes VanA and VanB, which confer high-level resistance to vancomycin in enterococci. This suggests that the resistance proteins currently found in clinical enterococcal isolates may have originated from glycopeptide-producing organisms. The G+C contents of ddlM and ddlN are high (65% and 58%, respectively) compared with VanA (45%) and VanB (48%). Thus recent horizontal transfer of the ddl genes directly from these drug-producing organisms is not evidenced. Nonetheless, the very high amino acid sequence homology suggests possible transfer from a related producer or perhaps via one or more bacterial intermediaries. That antibiotic-producing organisms are the source of resistance genes is not a new concept (30). More troubling, however, is recent evidence that has shown that some antibiotic preparations are, in fact, contaminated by DNA from producing organisms, including antibiotic-resistance genes (31). These results signal again that the wide spread use of members of groups of antibiotics with clinical importance should be closely monitored.
Acknowledgments
We thank Dr. Astrid Petrich and the members of Dr. R. Gupta’s group for helpful suggestions during gene cloning. A. orientalis C329.2 was the kind gift of Eli Lilly and Company. This work was funded by the Natural Sciences and Engineering Research Council of Canada and by a Medical Research Council of Canada Graduate Studentship to C.G.M.
ABBREVIATIONS
- VRE
vancomycin-resistant enterococci
- d-Lac
d-lactate
- Ddl
d-Ala-d-Ala ligase
- DdlM
Ddl from Streptomyces toyocaensis
- DdlN
Ddl from Amycolatopsis orientalis
Footnotes
References
- 1.Murray B E. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Courvalin P. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh C T, Fisher S L, Park I-S, Prohalad M, Wu Z. Chem Biol. 1996;3:21–28. doi: 10.1016/s1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]
- 4.Arthur M, Molinas C, Depardieu F, Courvalin P. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bugg T D H, Wright G D, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Mol Microbiol. 1994;13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z, Wright G D, Walsh C T. Biochemistry. 1995;34:2455–2463. doi: 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly J P, Voss A, Witte W, Murray B E. J Antimicrob Chemother. 1996;37:389–397. doi: 10.1093/jac/37.2.389. [DOI] [PubMed] [Google Scholar]
- 9.Hayes P, Gustafson R H, Lotgering I F K, Mudd A J. J Antimicrob Chemother. 1996;37:390–392. [Google Scholar]
- 10.Howarth F, Poulter D. Lancet. 1996;347:1047. doi: 10.1016/s0140-6736(96)90187-7. [DOI] [PubMed] [Google Scholar]
- 11.Mudd A. Lancet. 1996;347:1412. doi: 10.1016/s0140-6736(96)91055-7. [DOI] [PubMed] [Google Scholar]
- 12.Wise R. Lancet. 1996;347:1835. doi: 10.1016/s0140-6736(96)91654-2. [DOI] [PubMed] [Google Scholar]
- 13.Bates J, Jordens J Z, Griffiths D T. J Antimicrob Chemother. 1994;34:507–516. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 14.Klare I, Heier H, Claus R, Reissbrodt R, Witte W. FEMS Microbiol Lett. 1995;125:165–171. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 15.Torres C, Reguera J A, Sanmartin M J, Perez-Diaz J C, Baquero F. J Antimicrob Chemother. 1994;33:553–561. doi: 10.1093/jac/33.3.553. [DOI] [PubMed] [Google Scholar]
- 16.Coque T M, Tomayko J F, Ricke S C, Okhyusen P C, Murray B E. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutka-Malen S, Molinas C, Arthur M, Courvalin P. Mol Gen Genet. 1990;224:364–372. doi: 10.1007/BF00262430. [DOI] [PubMed] [Google Scholar]
- 18.Evers S, Reynolds P E, Courvalin P. Gene. 1994;140:97–102. doi: 10.1016/0378-1119(94)90737-4. [DOI] [PubMed] [Google Scholar]
- 19.Evers S, Casadewall B, Charles M, Dutka-Malen S, Galimand M, Courvalin P. J Mol Evol. 1996;42:706–712. doi: 10.1007/BF02338803. [DOI] [PubMed] [Google Scholar]
- 20.Dutka-Malen S, Molinas C, Arthur M, Courvalin P. Gene. 1992;112:53–58. doi: 10.1016/0378-1119(92)90302-6. [DOI] [PubMed] [Google Scholar]
- 21.Denis F, Brzezinski R. Gene. 1992;111:115–118. doi: 10.1016/0378-1119(92)90611-r. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Higgins D G, Sharp P M. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 24.Dayhoff M O. In: Atlas of Protein Sequence and Structure. Dayhoff M O, editor. Vol. 5. Washington, DC: National Biomedical Research Foundation; 1978. , Suppl. 3, pp. 345–358. [Google Scholar]
- 25.Fan C, Moews P C, Walsh C T, Knox J R. Science. 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Walsh C T. Biochemistry. 1995;34:2768–2776. doi: 10.1021/bi00009a005. [DOI] [PubMed] [Google Scholar]
- 27.Park I-S, Lin C-H, Walsh C T. Biochemistry. 1996;35:10464–10471. doi: 10.1021/bi9603128. [DOI] [PubMed] [Google Scholar]
- 28.Wright G D, Walsh C T. Protein Sci. 1993;2:1765–1769. doi: 10.1002/pro.5560021020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arthur M, Molinas C, Bugg T D H, Wright G D, Courvalin P. Antimicrob Agents Chemother. 1992;36:867–869. doi: 10.1128/aac.36.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benveniste R, Davies J. Proc Natl Acad Sci USA. 1973;70:2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb V, Davies J. Antimicrob Agents Chemother. 1993;37:2379–2384. doi: 10.1128/aac.37.11.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]