Abstract
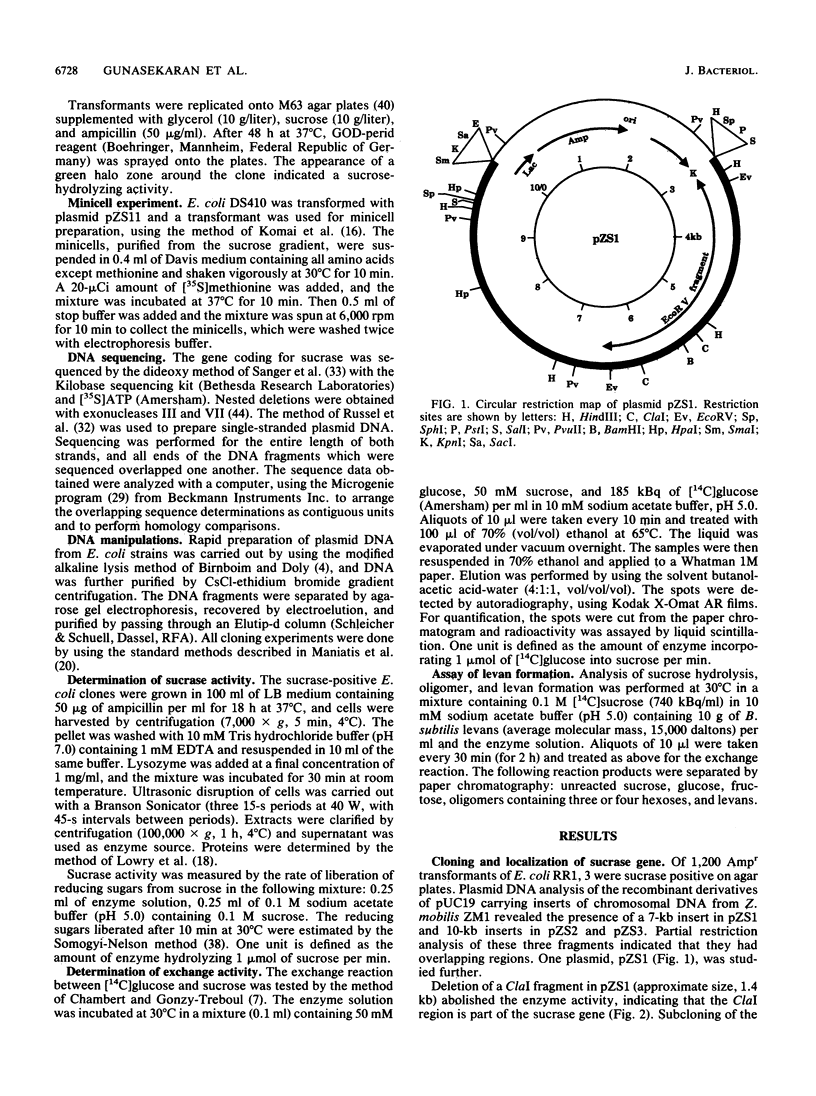
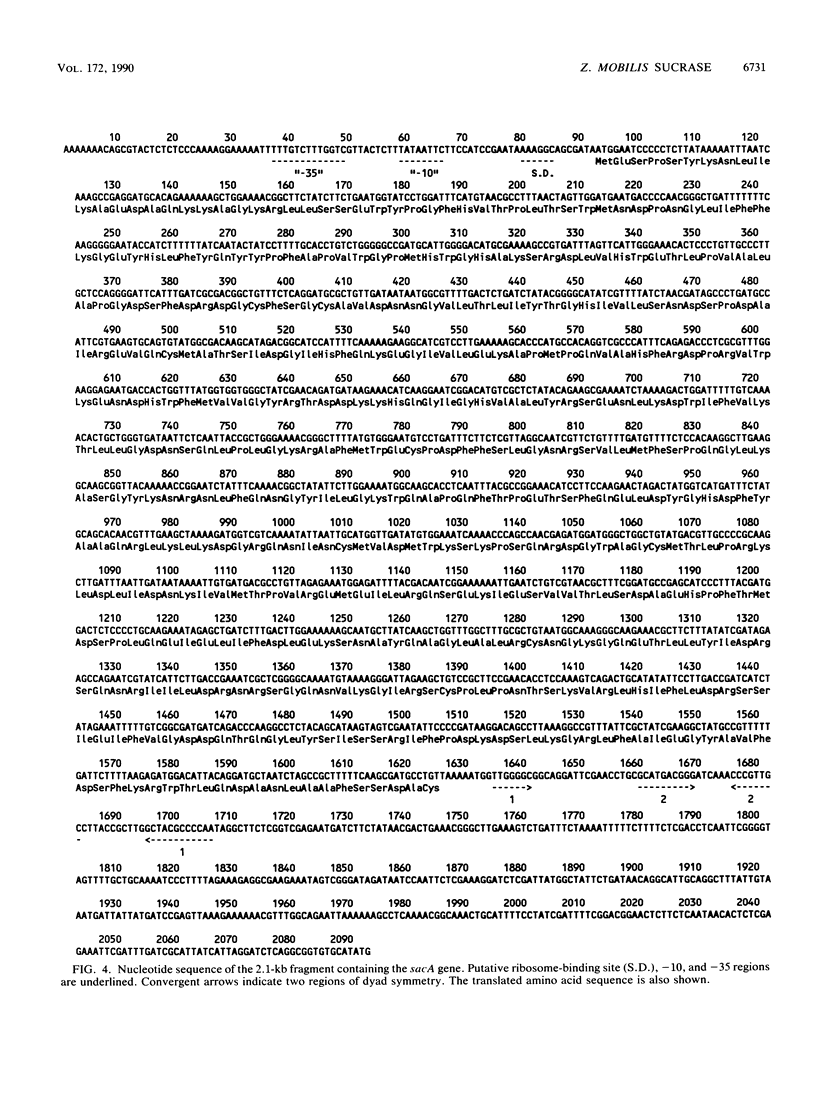
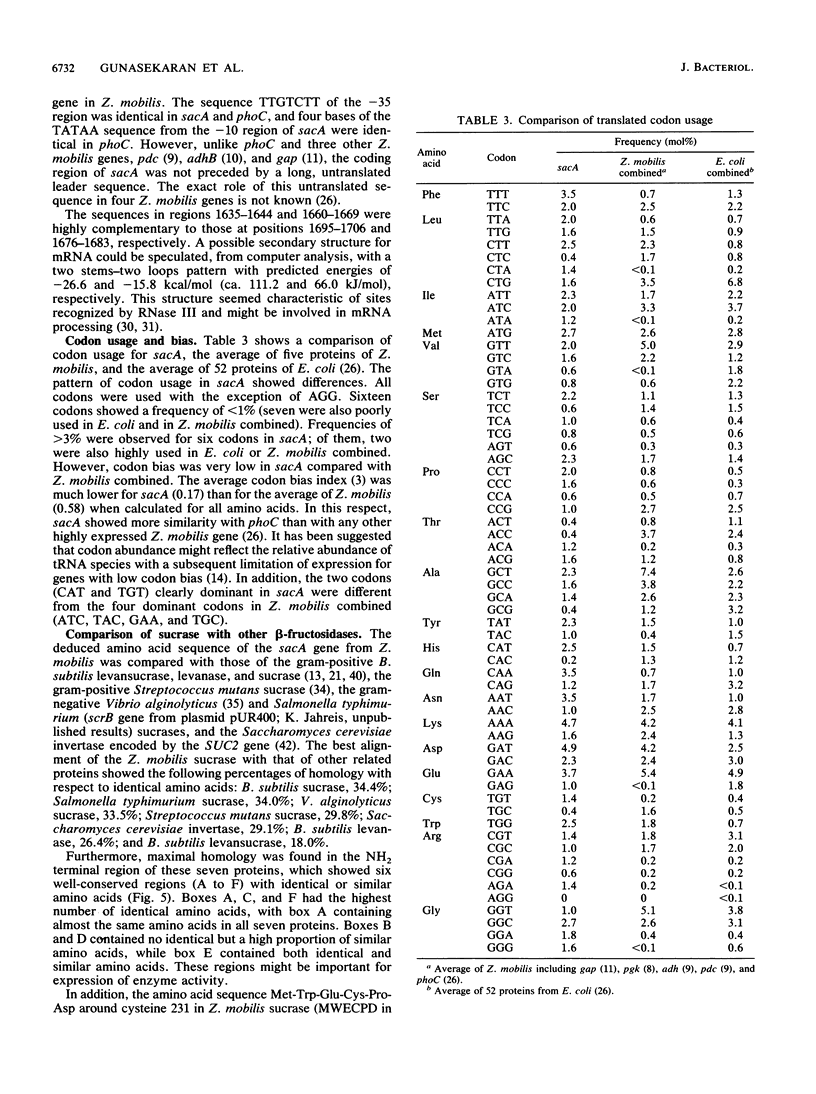
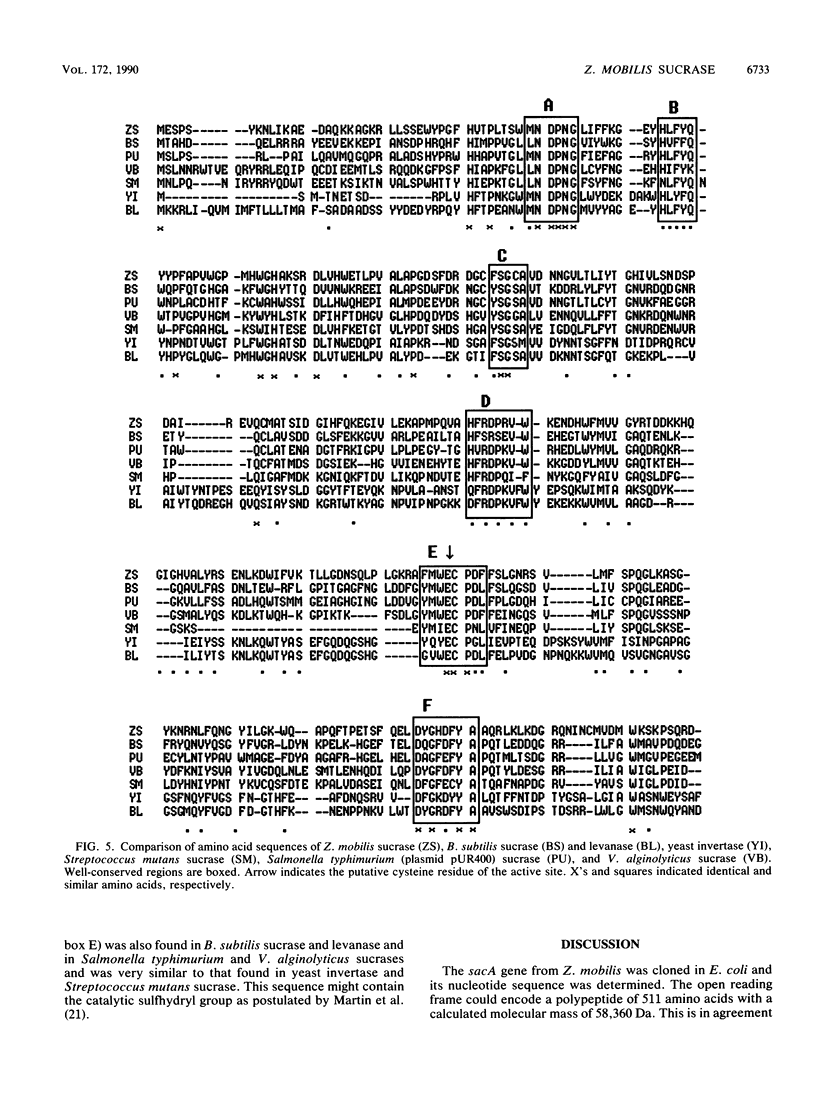
The Zymomonas mobilis gene (sacA) encoding a protein with sucrase activity has been cloned in Escherichia coli and its nucleotide sequence has been determined. Potential ribosome-binding site and promoter sequences were identified in the region upstream of the gene which were homologous to E. coli and Z. mobilis consensus sequences. Extracts from E. coli cells, containing the sacA gene, displayed a sucrose-hydrolyzing activity. However, no transfructosylation activity (exchange reaction or levan formation) could be detected. This sucrase activity was different from that observed with the purified extracellular protein B46 from Z. mobilis. These two proteins showed different electrophoretic mobilities and molecular masses and shared no immunological similarity. Thus, the product of sacA (a polypeptide of 58.4-kDa molecular mass) is a new sucrase from Z. mobilis. The amino acid sequence, deduced from the nucleotide sequence of sacA, showed strong homologies with the sucrases from Bacillus subtilis, Salmonella typhimurium, and Vibrio alginolyticus.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baratti J. C., Bu'lock J. D. Zymomonas mobilis: a bacterium for ethanol production. Biotechnol Adv. 1986;4(1):95–115. doi: 10.1016/0734-9750(86)90006-6. [DOI] [PubMed] [Google Scholar]
- Barrow K. D., Collins J. G., Rogers P. L., Smith G. M. The structure of a novel polysaccharide isolated from Zymomonas mobilis determined by nuclear magnetic resonance spectroscopy. Eur J Biochem. 1984 Nov 15;145(1):173–179. doi: 10.1111/j.1432-1033.1984.tb08537.x. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Chambert R., Gonzy-Tréboul G. Levansucrase of Bacillus subtilis: kinetic and thermodynamic aspects of transfructosylation processes. Eur J Biochem. 1976 Feb 2;62(1):55–64. doi: 10.1111/j.1432-1033.1976.tb10097.x. [DOI] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988 Apr;170(4):1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Ingram L. O. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J Bacteriol. 1987 Dec;169(12):5653–5662. doi: 10.1128/jb.169.12.5653-5662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimarco A. A., Romano A. H. d-Glucose Transport System of Zymomonas mobilis. Appl Environ Microbiol. 1985 Jan;49(1):151–157. doi: 10.1128/aem.49.1.151-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A., Klier A., Rapoport G. Nucleotide sequence of the sucrase gene of Bacillus subtilis. Gene. 1986;45(2):221–225. doi: 10.1016/0378-1119(86)90258-1. [DOI] [PubMed] [Google Scholar]
- Hoekema A., Kastelein R. A., Vasser M., de Boer H. A. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol Cell Biol. 1987 Aug;7(8):2914–2924. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klier A. F., Rapoport G. Genetics and regulation of carbohydrate catabolism in Bacillus. Annu Rev Microbiol. 1988;42:65–95. doi: 10.1146/annurev.mi.42.100188.000433. [DOI] [PubMed] [Google Scholar]
- Komai N., Nishizawa T., Hayakawa Y., Murotsu T., Matsubara K. Detection and mapping of six miniF-encoded proteins by cloning analysis of dissected miniF segments. Mol Gen Genet. 1982;186(2):193–203. doi: 10.1007/BF00331850. [DOI] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lefesant J. A., Walle J., Dedonder R. Purification and some properties of an endocellular sucrase from a constitutive mutant of Bacillus subtilis Marburg 168. Eur J Biochem. 1974 Mar 1;42(2):611–620. doi: 10.1111/j.1432-1033.1974.tb03376.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Martin I., Débarbouillé M., Ferrari E., Klier A., Rapoport G. Characterization of the levanase gene of Bacillus subtilis which shows homology to yeast invertase. Mol Gen Genet. 1987 Jun;208(1-2):177–184. doi: 10.1007/BF00330439. [DOI] [PubMed] [Google Scholar]
- Nossal N. G., Heppel L. A. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J Biol Chem. 1966 Jul 10;241(13):3055–3062. [PubMed] [Google Scholar]
- Pond J. L., Eddy C. K., Mackenzie K. F., Conway T., Borecky D. J., Ingram L. O. Cloning, sequencing, and characterization of the principal acid phosphatase, the phoC+ product, from Zymomonas mobilis. J Bacteriol. 1989 Feb;171(2):767–774. doi: 10.1128/jb.171.2.767-774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D. Escherichia coli ribonuclease III cleavage sites. Cell. 1982 Oct;30(3):669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
- Russel M., Kidd S., Kelley M. R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45(3):333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- Régnier P., Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986 Jan 5;187(1):23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- SMOGYI M. Notes on sugar determination. J Biol Chem. 1952 Mar;195(1):19–23. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Kuramitsu H. K. Sequence analysis of the Streptococcus mutans scrB gene. Infect Immun. 1988 Aug;56(8):1956–1960. doi: 10.1128/iai.56.8.1956-1960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle R. R., Robb S. M., Robb F. T., Woods D. R. Nucleotide sequence and analysis of the Vibrio alginolyticus sucrase gene (scrB). Gene. 1989 Aug 1;80(1):49–56. doi: 10.1016/0378-1119(89)90249-7. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Le Coq D., Aymerich S., Gonzy-Tréboul G., Gay P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet. 1985;200(2):220–228. doi: 10.1007/BF00425427. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Le Coq D., Djemia H. B., Gay P. Analyse génétique de sacB, gène de structure d'une enzyme secrétée, la lévane-saccharase de Bacillus subtilis Marburg. Mol Gen Genet. 1983;191(1):138–144. doi: 10.1007/BF00330901. [DOI] [PubMed] [Google Scholar]
- Swings J., De Ley J. The biology of Zymomonas. Bacteriol Rev. 1977 Mar;41(1):1–46. doi: 10.1128/br.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig R., Carlson M. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 1983 Mar 25;11(6):1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



