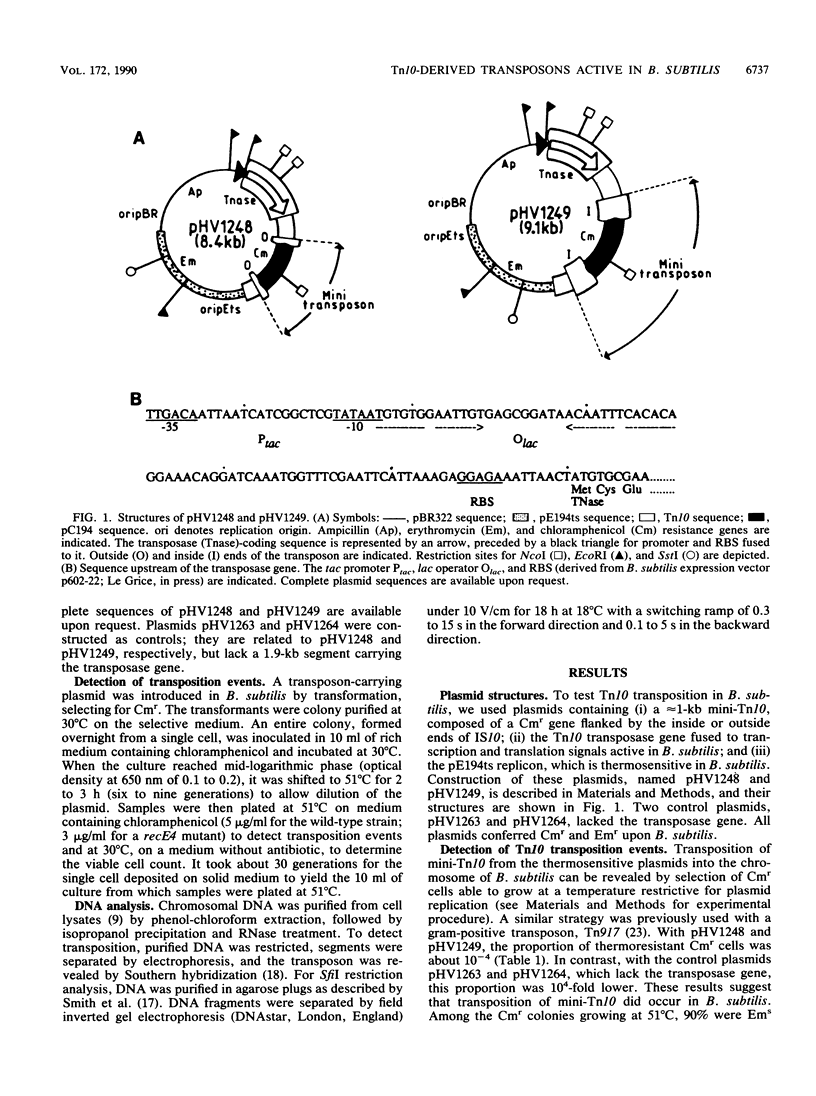
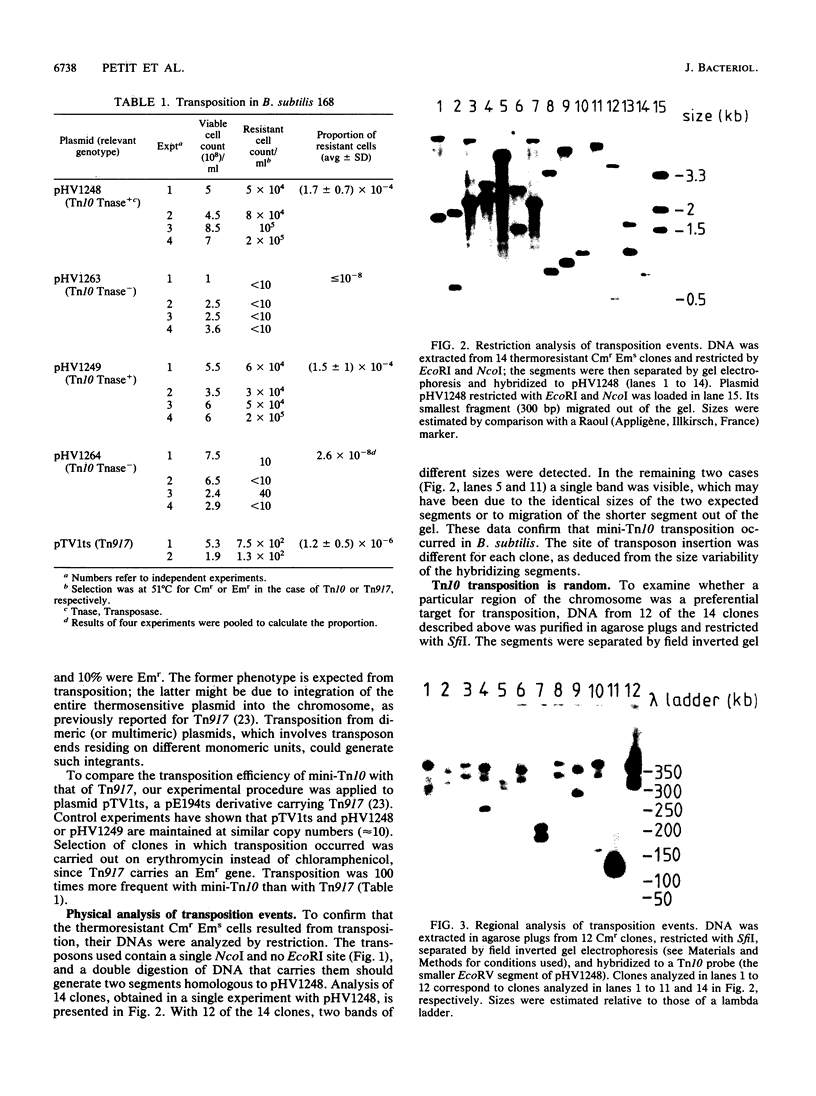
Abstract
Small derivatives of the Escherichia coli transposon Tn10, comprising IS10 ends and a chloramphenicol resistance gene, were introduced in Bacillus subtilis on a thermosensitive plasmid, pE194. In the presence of the Tn10 transposase gene fused to signals functional in B. subtilis, these derivatives transposed with a frequency of 10(-6) per element per generation. They had no highly preferred insertion site or region, as judged by restriction analysis of the chromosomal DNA, and generated auxotrophic and sporulation-deficient mutants with a frequency of about 1%. These results suggest that Tn10 derivatives might be a useful genetic tool in B. subtilis and possibly other gram-positive microorganisms.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Courvalin P., Carlier C. Tn1545: a conjugative shuttle transposon. Mol Gen Genet. 1987 Feb;206(2):259–264. doi: 10.1007/BF00333582. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. Insertion of foreign DNA into plasmids from gram-positive bacteria induces formation of high-molecular-weight plasmid multimers. J Bacteriol. 1988 Mar;170(3):1183–1190. doi: 10.1128/jb.170.3.1183-1190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Elkana Y., Ehrlich S. D., Lederberg J. Electrophoretic separation of Bacillus subtilis genes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2207–2211. doi: 10.1073/pnas.72.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordănescu S. Recombinant plasmid obtained from two different, compatible staphylococcal plasmids. J Bacteriol. 1975 Nov;124(2):597–601. doi: 10.1128/jb.124.2.597-601.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Steele D. A., Reichardt K., Botstein D. Specificity of insertion by the translocatable tetracycline-resistance element Tn10. Genetics. 1979 Aug;92(4):1023–1040. doi: 10.1093/genetics/92.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- Morisato D., Way J. C., Kim H. J., Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983 Mar;32(3):799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- Niaudet B., Ehrlich S. D. In vitro genetic labeling of Bacillus subtilis cryptic plasmid pHV400. Plasmid. 1979 Jan;2(1):48–58. doi: 10.1016/0147-619x(79)90005-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Sakaguchi K. Construction of a recombinant plasmid composed of B. subtilis leucine genes and a B. subtilis (natto) plasmid: its use as cloning vehicle in B. subtilis 168. Mol Gen Genet. 1978 Oct 24;165(3):269–276. doi: 10.1007/BF00332526. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P. J., Perkins J. B., Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]




