Abstract
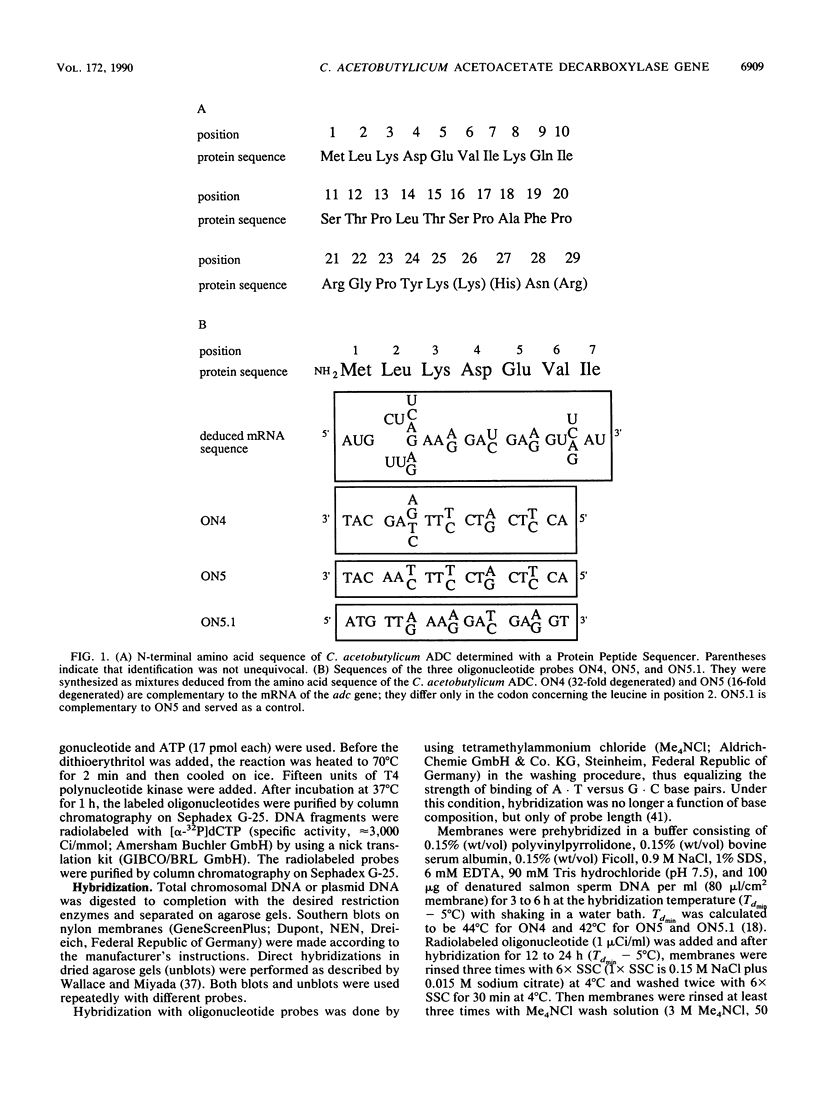
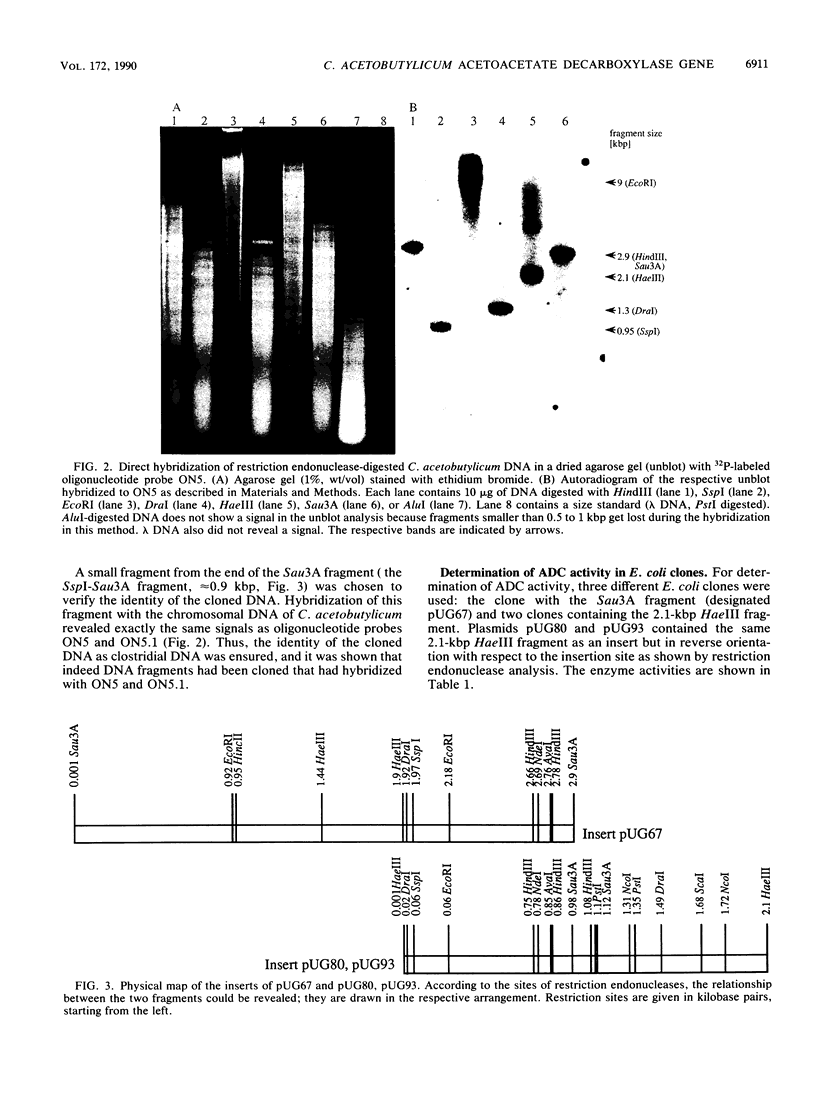
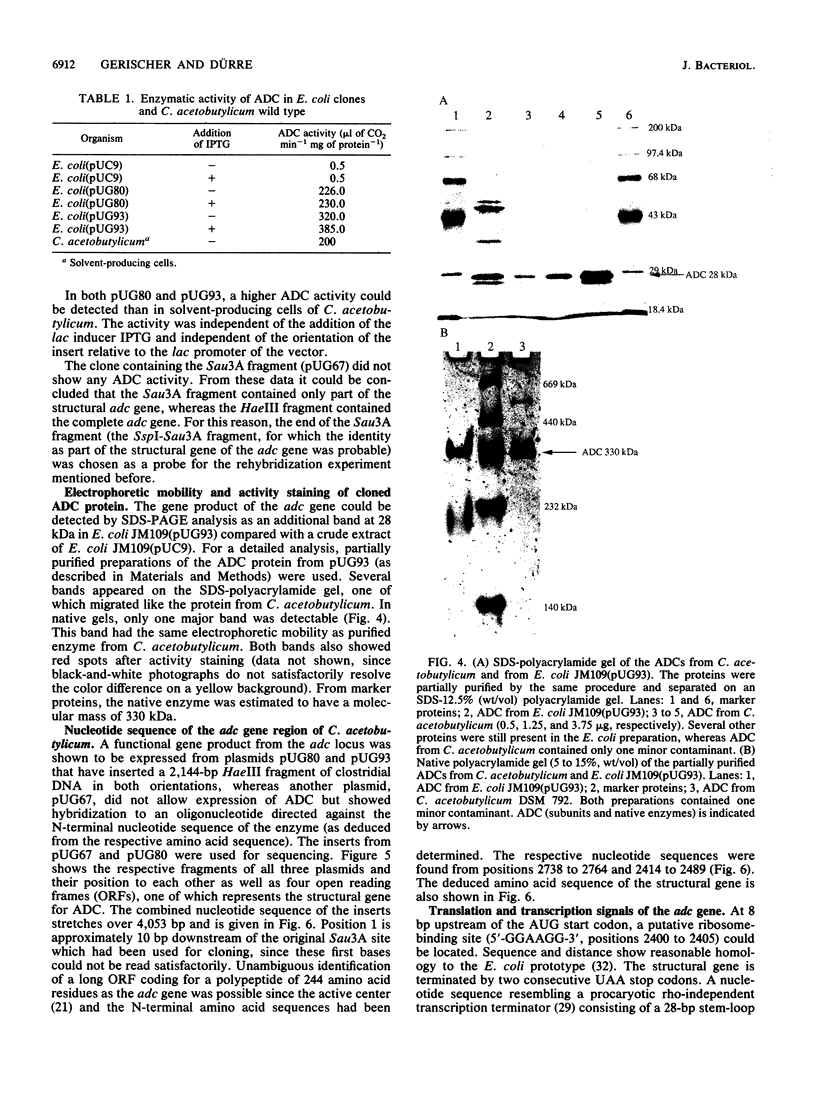
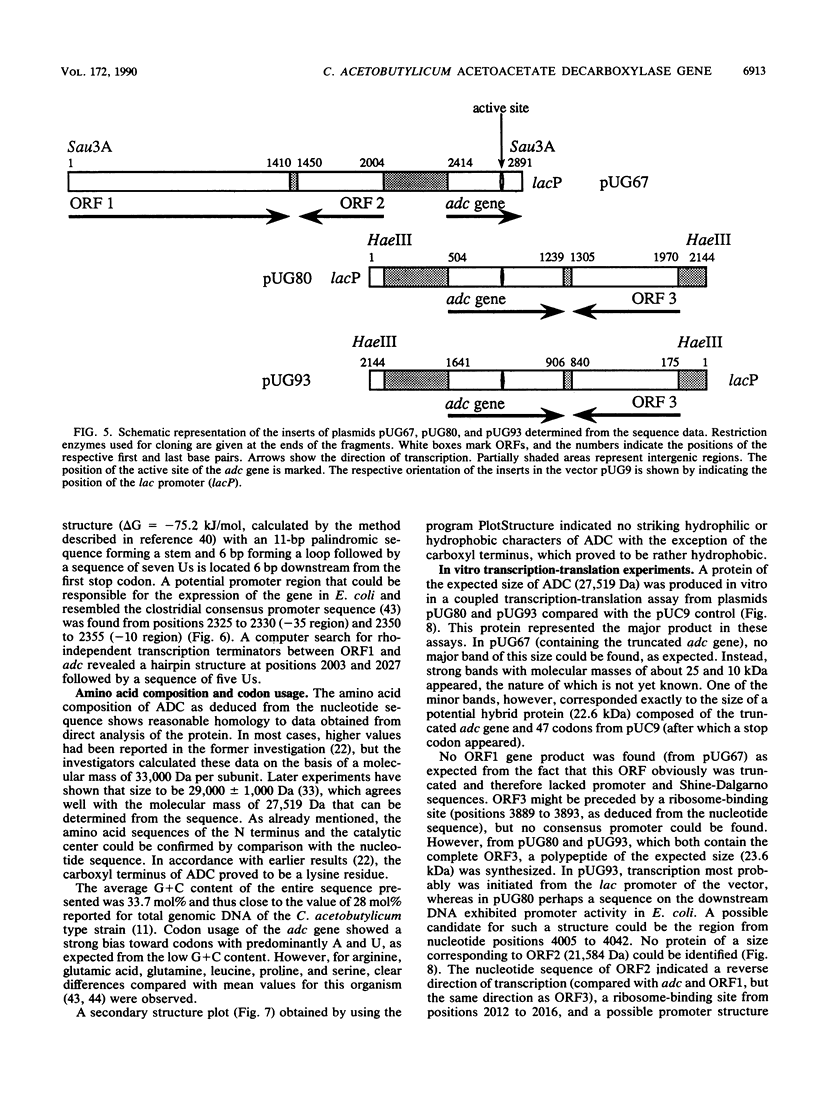
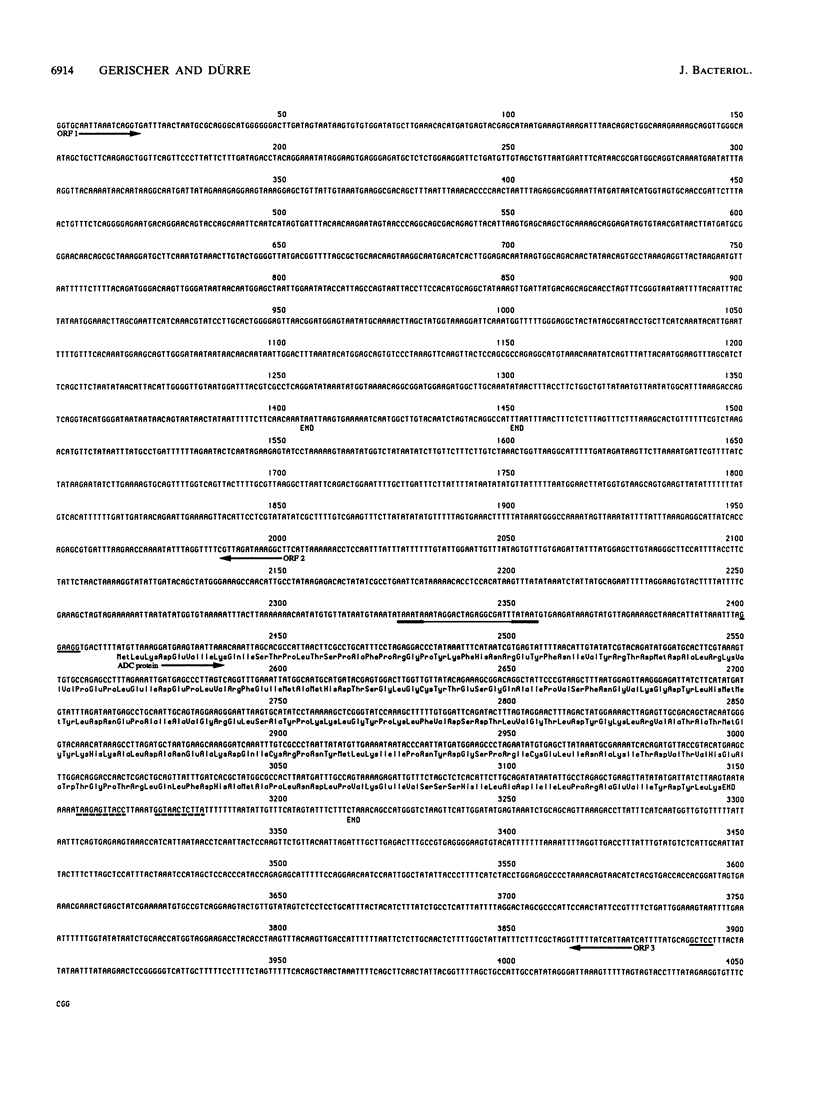
Acetoacetate decarboxylase (ADC) (EC4.1.1.4) of Clostridium acetobutylicum DSM 792 was purified to homogeneity, and its first 25 N-terminal amino acids were determined. Oligonucleotide probes deduced from this sequence were used to detect positive clones in partial gene banks derived from Sau3A and HaeIII digests with following ligation into the vector pUC9. In Escherichia coli, the 2.1-kbp HaeIII clones expressed high levels of ADC activity. The expression was independent of the orientation of the insert with respect to the lac promoter of the vector and also of the addition of isopropyl-beta-D-thiogalactopyranoside, thus indicating that sequences located on the clostridial DNA controlled transcription and translation. From the E. coli clone with the recombinant plasmid pUG93 containing the 2.1-kbp HaeIII fragment, the ADC protein was purified and compared with the native enzyme. Both were indistinguishable with respect to the molecular mass of subunits and native protein as well as to activity stain. The 2.9-kbp Sau3A fragment could be shown to contain the amino terminus of the acetoacetate decarboxylase (adc) gene but did not express enzyme activity. It partially overlapped with the HaeIII fragment, spanning together 4,053 bp of the clostridial genome that were completely sequenced. Four open reading frames (ORFs) could be detected, one of which was unambiguously assigned to the acetoacetate decarboxylase (adc) gene. Amino acid sequences of the N terminus and the catalytic center as deduced from the nucleotide sequence were identical to sequences obtained from direct analysis of the protein. Typical procaryotic transcriptional and translational start and stop signals could be found in the DNA sequence. Together with these regulatory sequences, the adc gene formed a single operon. The carboxyl terminus of the enzyme proved to be rather hydrophobic. In vitro transcription-translation assays resulted in formation of ADC and ORF3 gene product; the other two ORFs were not expressed. Whereas no homology of the adc gene and ORF2 could be detected with sequences available in the EMBL or GenBank data bases, the obviously truncated ORF1 showed significant similarity to alpha-amylase of Bacillus subtilis. The restriction pattern and N-terminal amino acid sequence (as deduced from the nucleotide sequence) of ORF3 proved to be identical to those of the large subunit of acetoacetyl coenzyme A:acetate/butyrate:coenzyme A transferase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P. J. Systems for polyacrylamide gel electrophoresis. Methods Enzymol. 1984;104:237–255. doi: 10.1016/s0076-6879(84)04093-3. [DOI] [PubMed] [Google Scholar]
- Cary J. W., Petersen D. J., Papoutsakis E. T., Bennett G. N. Cloning and expression of Clostridium acetobutylicum ATCC 824 acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase in Escherichia coli. Appl Environ Microbiol. 1990 Jun;56(6):1576–1583. doi: 10.1128/aem.56.6.1576-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIDOVICH I. LOCATION OF ACETOACETIC DECARBOXYLASE ON CELLULOSE ACETATE ELECTROPHORETOGRAMS BY MICRODIFFUSION OF PRODUCT. Anal Biochem. 1964 Mar;7:371–373. doi: 10.1016/0003-2697(64)90145-9. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Wallis J. Colony hybridization. Methods Enzymol. 1979;68:379–389. doi: 10.1016/0076-6879(79)68027-8. [DOI] [PubMed] [Google Scholar]
- Itakura K., Rossi J. J., Wallace R. B. Synthesis and use of synthetic oligonucleotides. Annu Rev Biochem. 1984;53:323–356. doi: 10.1146/annurev.bi.53.070184.001543. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Woods D. R. Acetone-butanol fermentation revisited. Microbiol Rev. 1986 Dec;50(4):484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laursen R. A., Westheimer F. H. The active site of cetoacetate decarboxylase. J Am Chem Soc. 1966 Jul 20;88(14):3426–3430. doi: 10.1021/ja00966a044. [DOI] [PubMed] [Google Scholar]
- Lederer F., Coutts S. M., Laursen R. A., Westheimer F. H. Acetoacetate decarboxylase. Subunits and properties. Biochemistry. 1966 Mar;5(3):823–833. doi: 10.1021/bi00867a003. [DOI] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather M. W. Base composition-independent hybridization in dried agarose gels: screening and recovery for cloning of genomic DNA fragments. Biotechniques. 1988 May;6(5):444–447. [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaki W., Westheimer F. H. Acetoacetate decarboxylase. The molecular weight of the enzyme and subunits. Biochemistry. 1968 Mar;7(3):895–900. doi: 10.1021/bi00843a002. [DOI] [PubMed] [Google Scholar]
- Verhasselt P., Poncelet F., Vits K., Van Gool A., Vanderleyden J. Cloning and expression of a Clostridium acetobutylicum alpha-amylase gene in Escherichia coli. FEMS Microbiol Lett. 1989 May;50(1-2):135–140. doi: 10.1111/j.1574-6968.1989.tb03097.x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vihinen M., Mäntsälä P. Microbial amylolytic enzymes. Crit Rev Biochem Mol Biol. 1989;24(4):329–418. doi: 10.3109/10409238909082556. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Miyada C. G. Oligonucleotide probes for the screening of recombinant DNA libraries. Methods Enzymol. 1987;152:432–442. doi: 10.1016/0076-6879(87)52050-x. [DOI] [PubMed] [Google Scholar]
- Wiesenborn D. P., Rudolph F. B., Papoutsakis E. T. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl Environ Microbiol. 1989 Feb;55(2):323–329. doi: 10.1128/aem.55.2.323-329.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young M., Minton N. P., Staudenbauer W. L. Recent advances in the genetics of the clostridia. FEMS Microbiol Rev. 1989 Dec;5(4):301–325. doi: 10.1111/j.1574-6968.1989.tb03402.x. [DOI] [PubMed] [Google Scholar]
- Youngleson J. S., Jones D. T., Woods D. R. Homology between hydroxybutyryl and hydroxyacyl coenzyme A dehydrogenase enzymes from Clostridium acetobutylicum fermentation and vertebrate fatty acid beta-oxidation pathways. J Bacteriol. 1989 Dec;171(12):6800–6807. doi: 10.1128/jb.171.12.6800-6807.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngleson J. S., Jones W. A., Jones D. T., Woods D. R. Molecular analysis and nucleotide sequence of the adh1 gene encoding an NADPH-dependent butanol dehydrogenase in the Gram-positive anaerobe Clostridium acetobutylicum. Gene. 1989 May 30;78(2):355–364. doi: 10.1016/0378-1119(89)90238-2. [DOI] [PubMed] [Google Scholar]
- Youngleson Jonathan S., Santangelo Joseph D., Jones David T., Woods David R. Cloning and Expression of a Clostridium acetobutylicum Alcohol Dehydrogenase Gene in Escherichia coli. Appl Environ Microbiol. 1988 Mar;54(3):676–682. doi: 10.1128/aem.54.3.676-682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerner B., Coutts S. M., Lederer F., Waters H. H., Westheimer F. H. Acetoacetate decarboxylase. Preparation of the enzyme. Biochemistry. 1966 Mar;5(3):813–816. doi: 10.1021/bi00867a001. [DOI] [PubMed] [Google Scholar]