Abstract
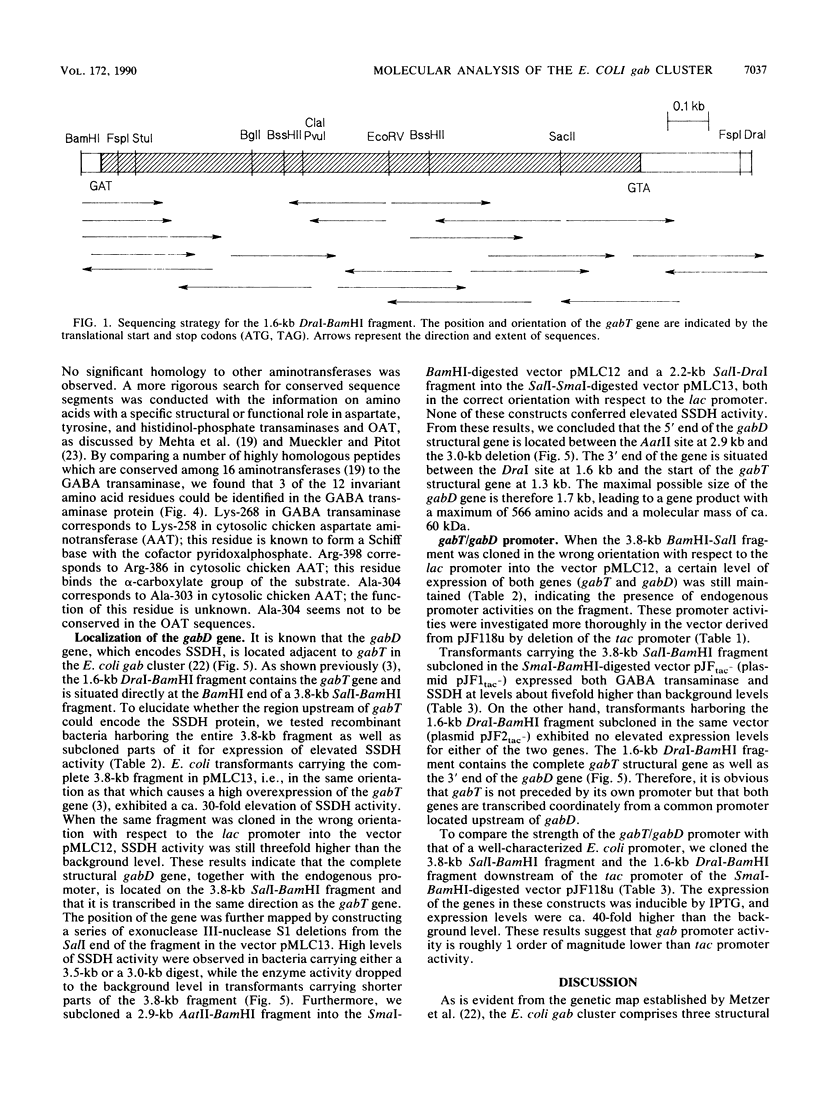
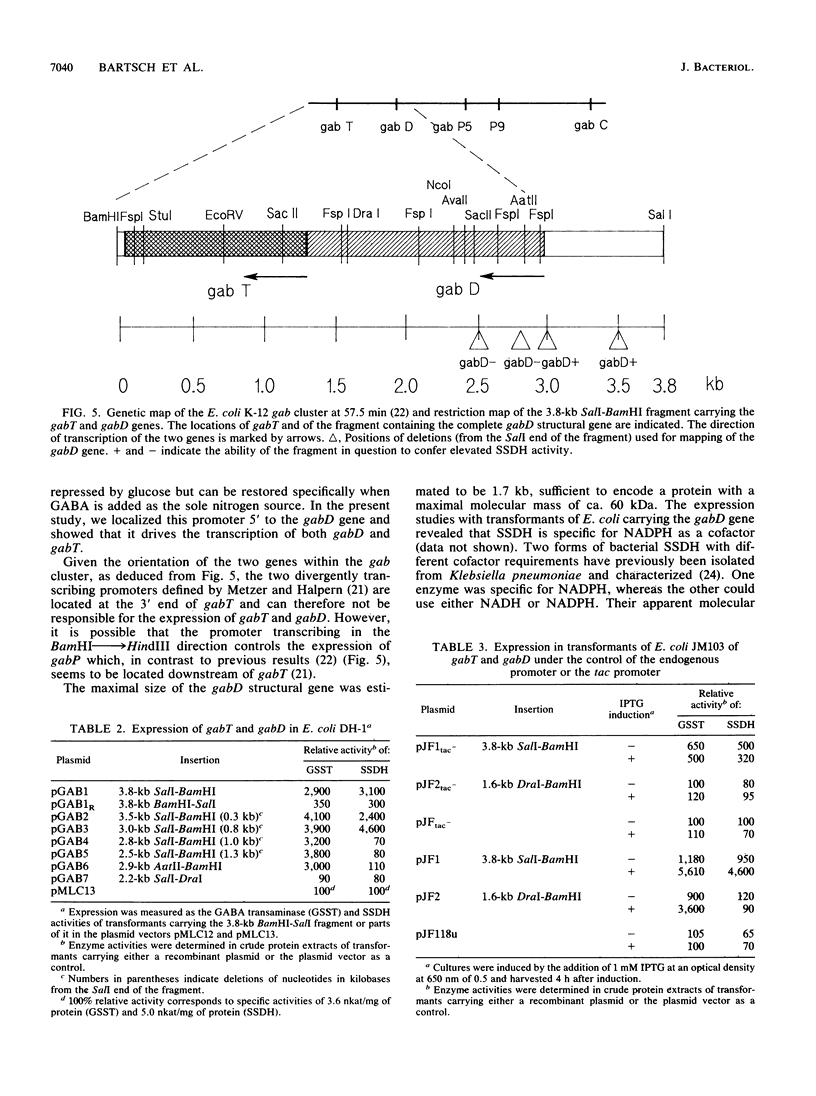
We have characterized two genes of the Escherichia coli K-12 gab cluster, which encodes the enzymes of the 4-aminobutyrate degradation pathway. The nucleotide sequence of gabT, coding for glutamate:succinic semialdehyde transaminase (EC 2.6.1.19), alternatively known as 4-aminobutyrate transaminase, was determined. The structural gene consists of 1,281 nucleotides specifying a protein of 426 amino acids with a molecular mass of 45.76 kDa. The protein shows significant homologies to the ornithine transaminases from Saccharomyces cerevisiae and from rat and human mitochondria. Three functionally and structurally important amino acid residues of the transaminase were identified by sequence comparison studies, and evolutionary relationships of the aminotransferases are discussed. The gabD gene, encoding succinic semialdehyde dehydrogenase (EC 1.2.1.16), was cloned and shown to be located adjacent to the 5' end of gabT. Expression studies with subfragments of the initially cloned DNA region revealed a maximal size of 1.7 kb for gabD. Both genes are cotranscribed from a promoter located upstream of gabD.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- André B., Jauniaux J. C. Nucleotide sequence of the yeast UGA1 gene encoding GABA transaminase. Nucleic Acids Res. 1990 May 25;18(10):3049–3049. doi: 10.1093/nar/18.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch K., Dichmann R., Schmitt P., Uhlmann E., Schulz A. Stereospecific production of the herbicide phosphinothricin (glufosinate) by transamination: cloning, characterization, and overexpression of the gene encoding a phosphinothricin-specific transaminase from Escherichia coli. Appl Environ Microbiol. 1990 Jan;56(1):7–12. doi: 10.1128/aem.56.1.7-12.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- De Biase D., Bolton J. B., Barra D., Bossa F., John R. A. Stoichiometry and stability of the adduct formed between human 4-aminobutyrate aminotransferase and 4-aminohex-5-enoate: sequence of a labelled peptide. Biochimie. 1989 Apr;71(4):491–495. doi: 10.1016/0300-9084(89)90179-x. [DOI] [PubMed] [Google Scholar]
- Dover S., Halpern Y. S. Control of the pathway of -aminobutyrate breakdown in Escherichia coli K-12. J Bacteriol. 1972 Apr;110(1):165–170. doi: 10.1128/jb.110.1.165-170.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover S., Halpern Y. S. Genetic analysis of the gamma-aminobutyrate utilization pathway in Escherichia coli K-12. J Bacteriol. 1974 Feb;117(2):494–501. doi: 10.1128/jb.117.2.494-501.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hsu L. C., Okamoto M., Snell E. E. L-Histidinol phosphate aminotransferase from Salmonella typhimurium. Kinetic behavior and sequence at the pyridoxal-P binding site. Biochimie. 1989 Apr;71(4):477–489. doi: 10.1016/0300-9084(89)90178-8. [DOI] [PubMed] [Google Scholar]
- Kim D. S., Churchich J. E. Active site modification of 4-aminobutyrate aminotransferase with ATP analogs. Biochim Biophys Acta. 1987 Dec 18;916(3):265–270. doi: 10.1016/0167-4838(87)90169-5. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Levinson A., Silver D., Seed B. Minimal size plasmids containing an M13 origin for production of single-strand transducing particles. J Mol Appl Genet. 1984;2(6):507–517. [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P. K., Hale T. I., Christen P. Evolutionary relationships among aminotransferases. Tyrosine aminotransferase, histidinol-phosphate aminotransferase, and aspartate aminotransferase are homologous proteins. Eur J Biochem. 1989 Dec 8;186(1-2):249–253. doi: 10.1111/j.1432-1033.1989.tb15202.x. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzer E., Halpern Y. S. In vivo cloning and characterization of the gabCTDP gene cluster of Escherichia coli K-12. J Bacteriol. 1990 Jun;172(6):3250–3256. doi: 10.1128/jb.172.6.3250-3256.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzer E., Levitz R., Halpern Y. S. Isolation and properties of Escherichia coli K-12 mutants impaired in the utilization of gamma-aminobutyrate. J Bacteriol. 1979 Mar;137(3):1111–1118. doi: 10.1128/jb.137.3.1111-1118.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M. M., Pitot H. C. Sequence of the precursor to rat ornithine aminotransferase deduced from a cDNA clone. J Biol Chem. 1985 Oct 25;260(24):12993–12997. [PubMed] [Google Scholar]
- Sanchez M., Fernández J., Martin M., Gibello A., Garrido-Pertierra A. Purification and properties of two succinic semialdehyde dehydrogenases from Klebsiella pneumoniae. Biochim Biophys Acta. 1989 Mar 24;990(3):225–231. doi: 10.1016/s0304-4165(89)80038-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A., Taggeselle P., Tripier D., Bartsch K. Stereospecific production of the herbicide phosphinothricin (glufosinate) by transamination: isolation and characterization of a phosphinothricin-specific transaminase from Escherichia coli. Appl Environ Microbiol. 1990 Jan;56(1):1–6. doi: 10.1128/aem.56.1.1-6.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa K., Asano S., Masu Y., Kuramitsu S., Kagamiyama H., Tanaka H., Soda K. The primary structure of thermostable D-amino acid aminotransferase from a thermophilic Bacillus species and its correlation with L-amino acid aminotransferases. J Biol Chem. 1989 Feb 15;264(5):2450–2454. [PubMed] [Google Scholar]
- Urdea M. S., Merryweather J. P., Mullenbach G. T., Coit D., Heberlein U., Valenzuela P., Barr P. J. Chemical synthesis of a gene for human epidermal growth factor urogastrone and its expression in yeast. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7461–7465. doi: 10.1073/pnas.80.24.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Sakabe K., Sakabe N., Higashi T., Sasaki K., Aibara S., Morita Y., Yonaha K., Toyama S., Fukutani H. Crystal structure analysis of omega-amino acid:pyruvate aminotransferase with a newly developed Weissenberg camera and an imaging plate using synchrotron radiation. J Biochem. 1989 Jan;105(1):1–3. doi: 10.1093/oxfordjournals.jbchem.a122600. [DOI] [PubMed] [Google Scholar]
- Yonaha K., Suzuki K., Toyama S. 4-Aminobutyrate:2-oxoglutarate aminotransferase of Streptomyces griseus: purification and properties. Eur J Biochem. 1985 Jan 2;146(1):101–106. doi: 10.1111/j.1432-1033.1985.tb08625.x. [DOI] [PubMed] [Google Scholar]
- Yonaha K., Toyama S. gamma-Aminobutyrate:alpha-ketoglutarate aminotransferase from Pseudomonas sp. F-126: purification, crystallization, and enzymologic properties. Arch Biochem Biophys. 1980 Mar;200(1):156–164. doi: 10.1016/0003-9861(80)90342-2. [DOI] [PubMed] [Google Scholar]



