Abstract
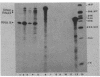
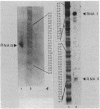
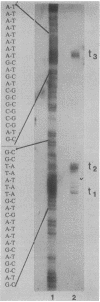
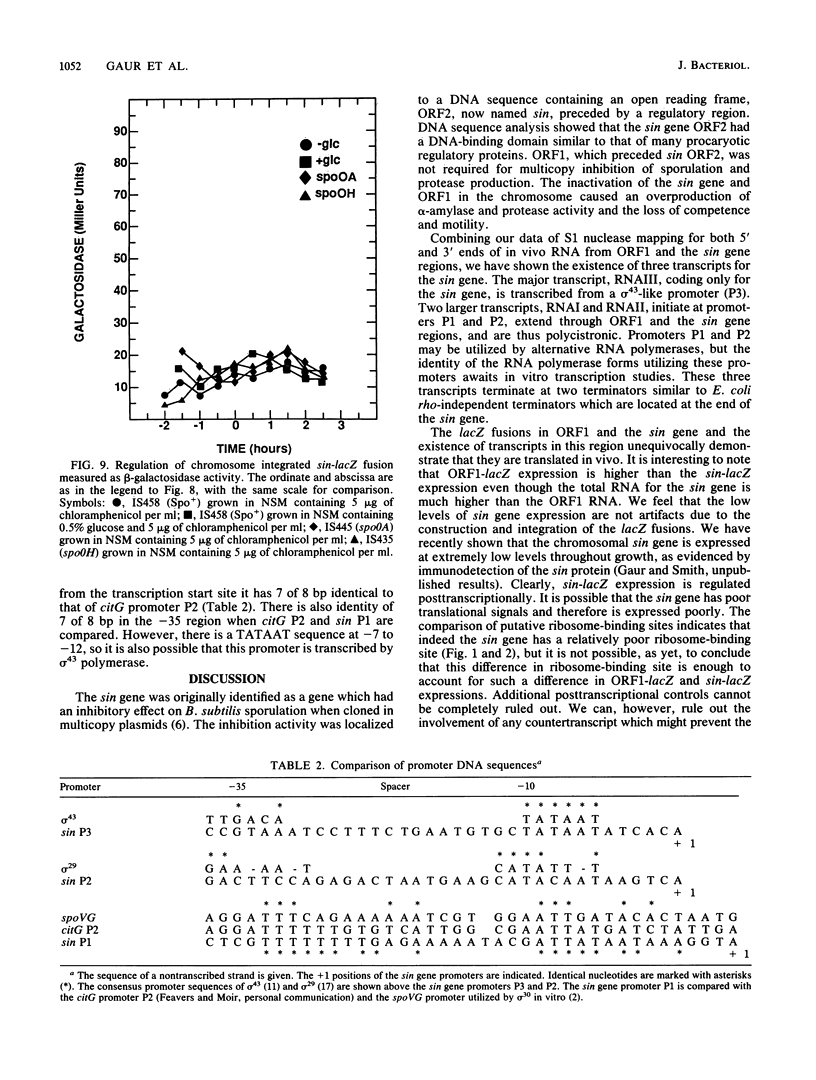
The newly identified sin gene affects late growth processes in Bacillus subtilis when it is overexpressed or inactivated in the chromosome. S1 nuclease mapping of the sin gene transcripts in vivo reveals the existence of three transcripts (RNAI, RNAII, and RNAIII). By correlating 5' ends of sin gene transcripts with DNA sequence, we have identified three different promoterlike sequences (P1, P2, and P3) for these transcripts. 3'-End mapping of these transcripts identified three prominent termination sites at the end of the sin gene. These termination sites are localized on two hairpin structures previously identified from the DNA sequence. The most abundant transcript, RNAIII, coded only for the sin gene, while the polycistronic transcripts RNAII and RNAI coded for the sin gene and ORF1 that precedes the sin gene. S1 mapping and translational lacZ fusion studies indicated that ORF1 and the sin gene are regulated differently. ORF1 expression is under developmental control, increasing at the end of vegetative growth, and requires functional spo0A and spo0H gene products. The sin gene is expressed at an almost constant and relatively low level throughout growth and remains largely unaffected by spo0A and spo0H mutations.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H. L., 3rd, Moran C. P., Jr New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9438–9442. doi: 10.1073/pnas.83.24.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E. J., Cabane K., Smith I. Regulation of spo0H, an early sporulation gene in bacilli. J Bacteriol. 1987 Mar;169(3):1182–1191. doi: 10.1128/jb.169.3.1182-1191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E., Weir J., Nair G., Carter L., 3rd, Moran C., Jr, Smith I. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol. 1988 Mar;170(3):1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., Hoch J. A. Sequence analysis of the spo0B locus reveals a polycistronic transcription unit. J Bacteriol. 1985 Feb;161(2):556–562. doi: 10.1128/jb.161.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Dubnau E., Smith I. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J Bacteriol. 1986 Nov;168(2):860–869. doi: 10.1128/jb.168.2.860-869.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. Genetics of bacterial sporulation. Adv Genet. 1976;18:69–98. doi: 10.1016/s0065-2660(08)60437-x. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Lewandoski M., Dubnau E., Smith I. Transcriptional regulation of the spo0F gene of Bacillus subtilis. J Bacteriol. 1986 Nov;168(2):870–877. doi: 10.1128/jb.168.2.870-877.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Maher D. L., Dennis P. P. In vivo transcription of E. coli genes coding for rRNA, ribosomal proteins and subunits of RNA polymerase: influence of the stringent control system. Mol Gen Genet. 1977 Oct 20;155(2):203–211. doi: 10.1007/BF00393161. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather P. N., Hay R. E., Ray G. L., Haldenwang W. G., Moran C. P., Jr Nucleotide sequences that define promoters that are used by Bacillus subtilis sigma-29 RNA polymerase. J Mol Biol. 1986 Dec 5;192(3):557–565. doi: 10.1016/0022-2836(86)90276-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. F., Doi R. H. Nucleotide sequence and organization of Bacillus subtilis RNA polymerase major sigma (sigma 43) operon. Nucleic Acids Res. 1986 May 27;14(10):4293–4307. doi: 10.1093/nar/14.10.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]