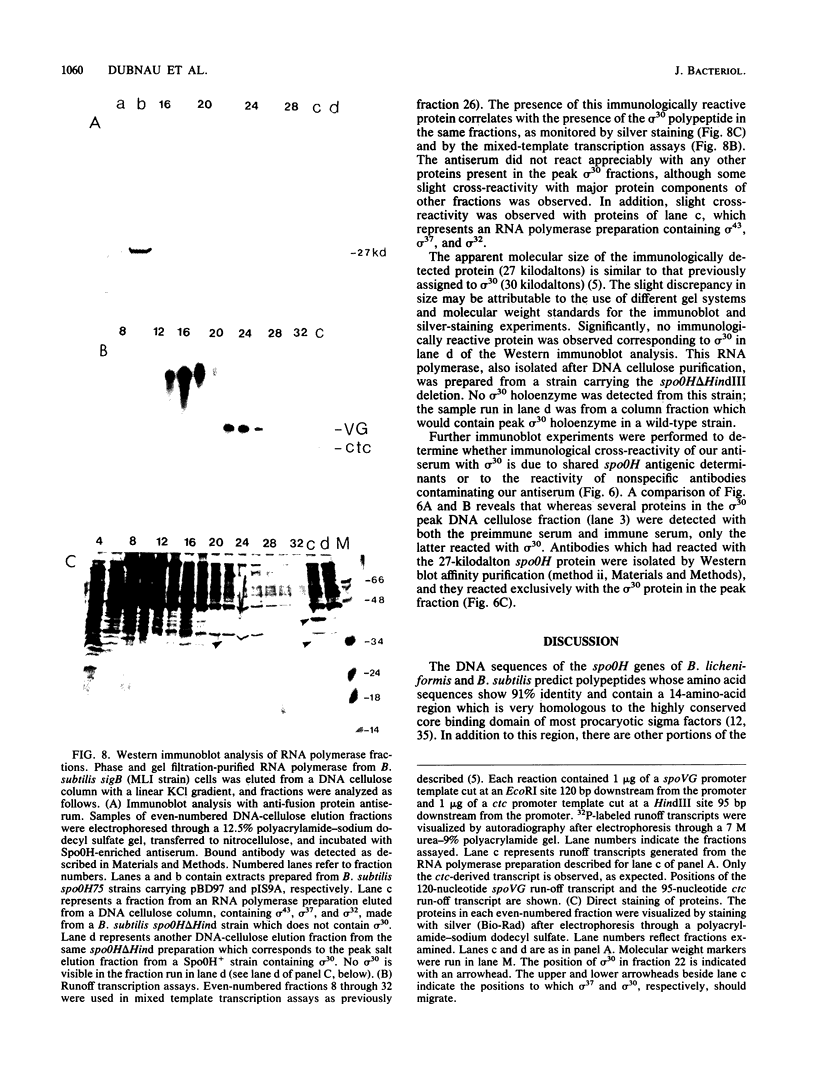
Abstract
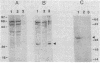
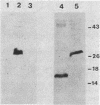
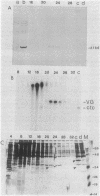
The DNA sequences of the spo0H genes from Bacillus licheniformis and B. subtilis are described, and the predicted open reading frames code for proteins of 26,097 and 25,447 daltons, respectively. The two spo0H gene products are 91% identical to one another and about 25% identical to most of the procaryotic sigma factors. The predicted proteins have a conserved 14-amino-acid sequence at their amino terminal end, typical of sigma factors. Antibodies raised against the spo0H gene product of B. licheniformis specifically react with RNA polymerase sigma factor protein, sigma 30, purified from B. subtilis. We conclude that the spo0H genes of B. licheniformis and B. subtilis code for sigma 30, now known as sigma H.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Hahn J., Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnie C., Lampe M., Losick R. Gene encoding the sigma 37 species of RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Bouvier J., Stragier P., Bonamy C., Szulmajster J. Nucleotide sequence of the spo0B gene of Bacillus subtilis and regulation of its expression. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7012–7016. doi: 10.1073/pnas.81.22.7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H. L., 3rd, Moran C. P., Jr New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9438–9442. doi: 10.1073/pnas.83.24.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Dubnau E. J., Cabane K., Smith I. Regulation of spo0H, an early sporulation gene in bacilli. J Bacteriol. 1987 Mar;169(3):1182–1191. doi: 10.1128/jb.169.3.1182-1191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E., Ramakrishna N., Cabane K., Smith I. Cloning of an early sporulation gene in Bacillus subtilis. J Bacteriol. 1981 Aug;147(2):622–632. doi: 10.1128/jb.147.2.622-632.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., Howard S. M., Hoch J. A. Effect of stage 0 sporulation mutations on subtilisin expression. J Bacteriol. 1986 Apr;166(1):173–179. doi: 10.1128/jb.166.1.173-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., Hoch J. A. Sequence analysis of the spo0B locus reveals a polycistronic transcription unit. J Bacteriol. 1985 Feb;161(2):556–562. doi: 10.1128/jb.161.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. D., Straus D. B., Walter W. A., Gross C. A. Sigma 32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1987 Apr;1(2):179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Hahn J., Contente S., Dubnau D. Replication and incompatibility properties of plasmid pE194 in Bacillus subtilis. J Bacteriol. 1982 Nov;152(2):722–735. doi: 10.1128/jb.152.2.722-735.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Israeli-Reches M., Dubnau D. Induction of macrolide-lincosamide-streptogramin B resistance requires ribosomes able to bind inducer. Mol Gen Genet. 1984;194(3):357–361. doi: 10.1007/BF00425544. [DOI] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Mathews J. L. Chromosomal location of pleiotropic negative sporulation mutations in Bacillus subtilis. Genetics. 1973 Feb;73(2):215–228. doi: 10.1093/genetics/73.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. P., Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi T., Kudoh J., Tsunasawa S. Amino-terminal structure of spoOA protein and sequence homology with spoOF and spoOB proteins. Mol Gen Genet. 1986 Jun;203(3):371–376. doi: 10.1007/BF00422059. [DOI] [PubMed] [Google Scholar]
- Ikeuchi T., Tsunasawa S., Sakiyama F. Purification and characterization of the spoOA protein of Bacillus subtilis from an overproducing strain of Escherichia coli. Eur J Biochem. 1987 Sep 1;167(2):233–238. doi: 10.1111/j.1432-1033.1987.tb13328.x. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., Banner C. D., Haldenwang W. G., Losick R. Promoter for a developmentally regulated gene in Bacillus subtilis. Cell. 1981 Sep;25(3):783–791. doi: 10.1016/0092-8674(81)90186-0. [DOI] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollington J. F., Haldenwang W. G., Huynh T. V., Losick R. Developmentally regulated transcription in a cloned segment of the Bacillus subtilis chromosome. J Bacteriol. 1981 Aug;147(2):432–442. doi: 10.1128/jb.147.2.432-442.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted J. B. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981 Dec 10;256(23):11955–11957. [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Perego M., Hoch J. A. Isolation and sequence of the spo0E gene: its role in initiation of sporulation in Bacillus subtilis. Mol Microbiol. 1987 Jul;1(1):125–132. doi: 10.1111/j.1365-2958.1987.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Piggot P. J. Mapping of asporogenous mutations of Bacillus subtilis: a minimum estimate of the number of sporeulation operons. J Bacteriol. 1973 Jun;114(3):1241–1253. doi: 10.1128/jb.114.3.1241-1253.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. W., Gitt M. A., Doi R. H. Isolation and physical mapping of the gene encoding the major sigma factor of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4074–4078. doi: 10.1073/pnas.80.13.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna N., Dubnau E., Smith I. The complete DNA sequence and regulatory regions of the Bacillus licheniformis spoOH gene. Nucleic Acids Res. 1984 Feb 24;12(4):1779–1790. doi: 10.1093/nar/12.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Parsot C., Bouvier J. Two functional domains conserved in major and alternate bacterial sigma factors. FEBS Lett. 1985 Jul 22;187(1):11–15. doi: 10.1016/0014-5793(85)81203-5. [DOI] [PubMed] [Google Scholar]
- Tatti K. M., Moran C. P., Jr Utilization of one promoter by two forms of RNA polymerase from Bacillus subtilis. Nature. 1985 Mar 14;314(6007):190–192. doi: 10.1038/314190a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K. A., Chapman J. W., Piggot P. J., Hoch J. A. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J., Chen S. M., Hoch J. A. Genetic analysis of a class of polymyxin resistant partial revertants of stage O sporulation mutants of Bacillus subtilis: map of the chromosome region near the origin of replication. Mol Gen Genet. 1979 May 23;173(1):61–70. doi: 10.1007/BF00267691. [DOI] [PubMed] [Google Scholar]
- Weir J., Dubnau E., Ramakrishna N., Smith I. Bacillus subtilis spo0H gene. J Bacteriol. 1984 Feb;157(2):405–412. doi: 10.1128/jb.157.2.405-412.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Ebert P. R., Stachel S. E., Gordon M. P., Nester E. W. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Yoshikawa H., Kawamura F., Takahashi H., Yamamoto T., Kobayashi Y., Saito H. The effect of spo0 mutations on the expression of spo0A- and spo0F-lacZ fusions. Mol Gen Genet. 1986 Oct;205(1):28–33. doi: 10.1007/BF02428029. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Kazami J., Yamashita S., Chibazakura T., Sone H., Kawamura F., Oda M., Isaka M., Kobayashi Y., Saito H. Revised assignment for the Bacillus subtilis spo0F gene and its homology with spo0A and with two Escherichia coli genes. Nucleic Acids Res. 1986 Jan 24;14(2):1063–1072. doi: 10.1093/nar/14.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]