Abstract
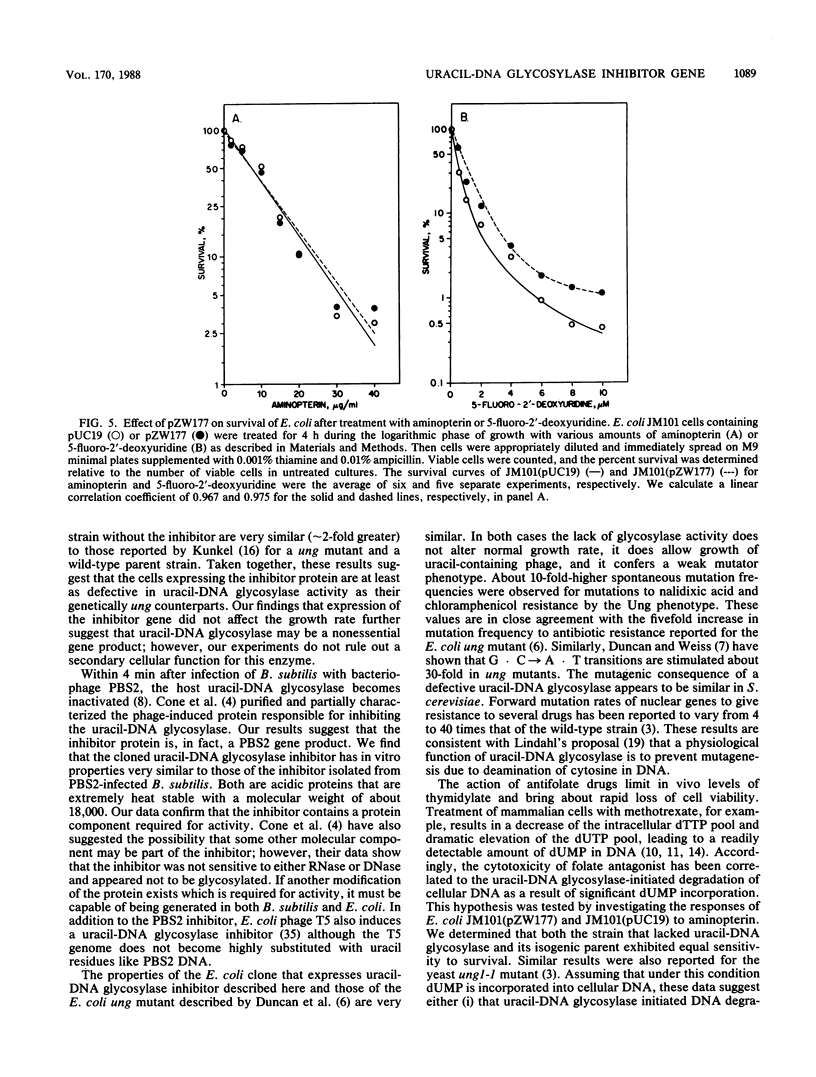
The uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 was cloned, and the effects of this inhibitor on Escherichia coli cells that contain uracil-DNA glycosylase activity were determined. A PBS2 genomic library was constructed by inserting EcoRI restriction fragments of PBS2 DNA into a plasmid pUC19 vector. The library was used to transform wild-type (ung+) E. coli, and the presence of the functional inhibitor gene was determined by screening for colonies that supported growth of M13mp19 phage containing uracil-DNA. A clone was identified that carried a 4.1-kilobase EcoRI DNA insert in the vector plasmid. Extracts of cells transformed with this recombinant plasmid lacked detectable uracil-DNA glycosylase activity and contained a protein that inhibited the activity of purified E. coli uracil-DNA glycosylase in vitro. The uracil-DNA glycosylase inhibitor expressed in these E. coli was partially purified and characterized as a heat-stable protein with a native molecular weight of about 18,000. Hence, we conclude that the PBS2 uracil-DNA glycosylase inhibitor gene was cloned and that the gene product has properties similar to those from PBS2-infected Bacillus subtilis cells. Inhibitor gene expression in E. coli resulted in (i) a weak mutator phenotype, (ii) a growth rate similar to that of E. coli containing pUC19 alone, (iii) a sensitivity to the antifolate drug aminopterin similar to that of cells lacking the inhibitor gene, and (iv) an increased resistance to the lethal effects of 5-fluoro-2'-deoxyuridine. These physiological properties are consistent with the phenotypes of other ung mutants.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Klein M. B. Selection by genetic transformation of a Saccharomyces cerevisiae mutant defective for the nuclear uracil-DNA-glycosylase. J Bacteriol. 1986 Jun;166(3):905–913. doi: 10.1128/jb.166.3.905-913.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone R., Bonura T., Friedberg E. C. Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2. Purification and preliminary characterization. J Biol Chem. 1980 Nov 10;255(21):10354–10358. [PubMed] [Google Scholar]
- Domena J. D., Mosbaugh D. W. Purification of nuclear and mitochondrial uracil-DNA glycosylase from rat liver. Identification of two distinct subcellular forms. Biochemistry. 1985 Dec 3;24(25):7320–7328. doi: 10.1021/bi00346a045. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Rockstroh P. A., Warner H. R. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978 Jun;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Ganesan A. K., Minton K. N-Glycosidase activity in extracts of Bacillus subtilis and its inhibition after infection with bacteriophage PBS2. J Virol. 1975 Aug;16(2):315–321. doi: 10.1128/jvi.16.2.315-321.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates F. T., 3rd, Linn S. Endonuclease V of Escherichia coli. J Biol Chem. 1977 Mar 10;252(5):1647–1653. [PubMed] [Google Scholar]
- Goulian M., Bleile B., Tseng B. Y. The effect of methotrexate on levels of dUTP in animal cells. J Biol Chem. 1980 Nov 25;255(22):10630–10637. [PubMed] [Google Scholar]
- Grafstrom R. H., Tseng B. Y., Goulian M. The incorporation of uracil into animal cell DNA in vitro. Cell. 1978 Sep;15(1):131–140. doi: 10.1016/0092-8674(78)90089-2. [DOI] [PubMed] [Google Scholar]
- Hitzeman R. A., Price A. R. Bacillus subtilis bacteriophage PBS2-induced DNA polymerase. Its purification and assay characteristics. J Biol Chem. 1978 Dec 10;253(23):8518–8525. [PubMed] [Google Scholar]
- Hochhauser S. J., Weiss B. Escherichia coli mutants deficient in deoxyuridine triphosphatase. J Bacteriol. 1978 Apr;134(1):157–166. doi: 10.1128/jb.134.1.157-166.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham H. A., Dickey L., Goulian M. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry. 1986 Jun 3;25(11):3225–3230. doi: 10.1021/bi00359a022. [DOI] [PubMed] [Google Scholar]
- Isolation and characterization of a Bacillus subtilis mutant with a defective N-glycosidase activity for uracil-containing deoxyribonucleic acid. J Bacteriol. 1977 Aug;131(2):438–445. doi: 10.1128/jb.131.2.438-445.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Cone R., Friedberg E. C. Specificity of the bacteriophage PBS2 induced inhibitor of uracil-DNA glycosylase. Biochemistry. 1981 Oct 13;20(21):6092–6096. doi: 10.1021/bi00524a027. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Li J. C., Kaminskas E. Accumulation of DNA strand breaks and methotrexate cytotoxicity. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5694–5698. doi: 10.1073/pnas.81.18.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Makino F., Munakata N. Deoxyuridine residues in DNA of thymine-requiring Bacillus subtilis strains with defective N-glycosidase activity for uracil-containing DNA. J Bacteriol. 1978 Apr;134(1):24–29. doi: 10.1128/jb.134.1.24-29.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. R., Cook S. J. New deoxyribonucleic acid polymerase induced by Bacillus subtilis bacteriophage PBS2. J Virol. 1972 Apr;9(4):602–610. doi: 10.1128/jvi.9.4.602-610.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. R., Fogt S. M. Deoxythymidylate phosphohydrolase induced by bacteriophage PBS2 during infection of Bacillus subtilis. J Biol Chem. 1973 Feb 25;248(4):1372–1380. [PubMed] [Google Scholar]
- Price A. R., Frato J. Bacillus subtilis deoxyuridinetriphosphatase and its bacteriophage PBS2-induced inhibitor. J Biol Chem. 1975 Nov 25;250(22):8804–8811. [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. Deoxyuridine triphosphatase of Escherichia coli. Purification, properties, and use as a reagent to reduce uracil incorporation into DNA. J Biol Chem. 1978 May 10;253(9):3305–3312. [PubMed] [Google Scholar]
- TAKAHASHI I., MARMUR J. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature. 1963 Feb 23;197:794–795. doi: 10.1038/197794a0. [DOI] [PubMed] [Google Scholar]
- Taylor A. F., Weiss B. Role of exonuclease III in the base excision repair of uracil-containing DNA. J Bacteriol. 1982 Jul;151(1):351–357. doi: 10.1128/jb.151.1.351-357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita F., Takahashi I. A novel enzyme, dCTP deaminase, found in Bacillus subtilis infected with phage PBS I. Biochim Biophys Acta. 1969 Mar 18;179(1):18–27. doi: 10.1016/0005-2787(69)90117-8. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Chien J., Lehman I. R., Duncan B. K., Warner H. R. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1978 Jan;75(1):233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B. K., Nyman P. O., Lehman I. R., Hochhauser S., Weiss B. Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc Natl Acad Sci U S A. 1977 Jan;74(1):154–157. doi: 10.1073/pnas.74.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Johnson L. K., Snustad D. P. Early events after infection of Escherichia coli by bacteriophage T5. III. Inhibition of uracil-DNA glycosylase activity. J Virol. 1980 Jan;33(1):535–538. doi: 10.1128/jvi.33.1.535-538.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., el-Hajj H. H. The repair of uracil-containing DNA. Basic Life Sci. 1986;38:349–356. doi: 10.1007/978-1-4615-9462-8_38. [DOI] [PubMed] [Google Scholar]
- Yoshioka A., Tanaka S., Hiraoka O., Koyama Y., Hirota Y., Ayusawa D., Seno T., Garrett C., Wataya Y. Deoxyribonucleoside triphosphate imbalance. 5-Fluorodeoxyuridine-induced DNA double strand breaks in mouse FM3A cells and the mechanism of cell death. J Biol Chem. 1987 Jun 15;262(17):8235–8241. [PubMed] [Google Scholar]