Abstract
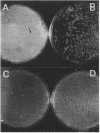
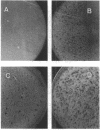
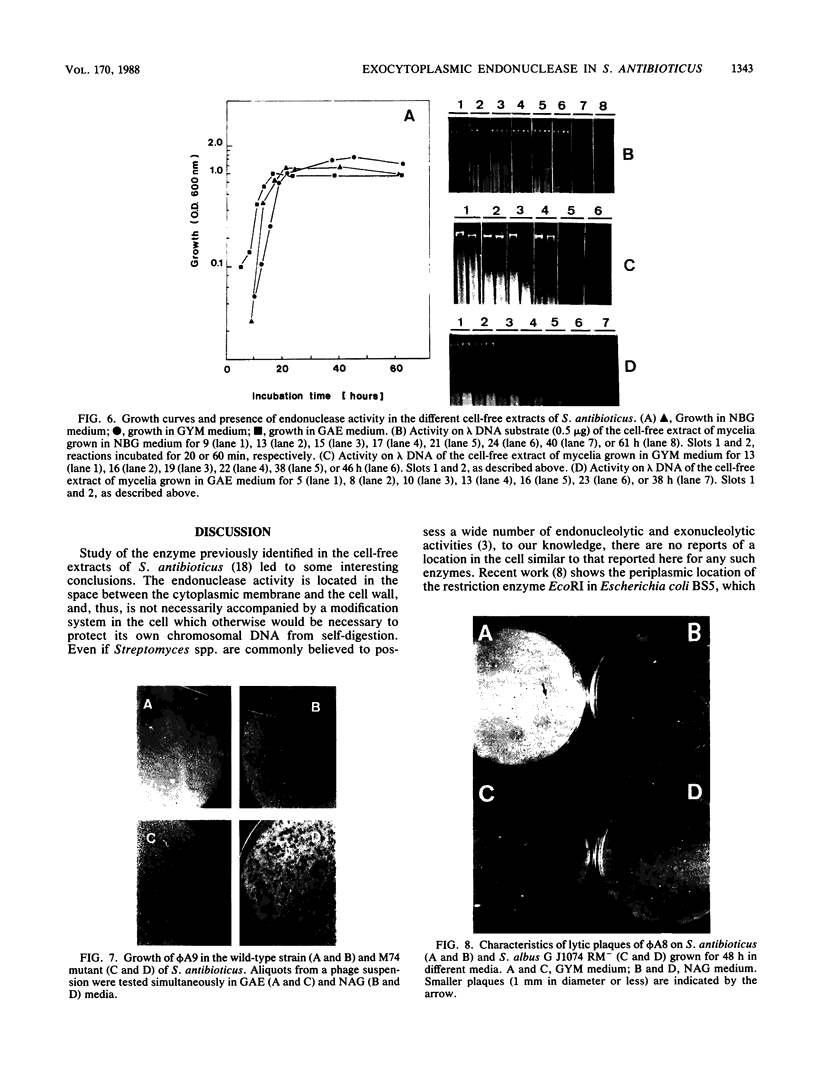
Streptomyces antibioticus produces a strong endo-DNase which is located between the cytoplasmic membrane and the cell wall. All DNA substrates assayed, including the chromosomal DNA of this species and several bacteriophage DNAs, were completely degraded in vitro by the enzyme. The rate of synthesis of the nuclease depended on the growth medium. In NBG medium, in which the enzyme is not produced, the size of lytic plaques of several actinophages was larger than that in GYM or GAE medium, in which synthesis of the nuclease takes place late in growth. In addition, one of the phages assayed, phi A6, showed a diminution of its efficiency of plating in GYM medium with respect to that in NBG medium; another phage, phi A9, grew in NBG medium but not in the other two media. It is postulated that the presence of the host nuclease, together with the capability of the particular phage to absorb on S. antibioticus of different growth phases, determines the efficiency of growth and the plaque size of the phages on productive media. This hypothesis was confirmed when the growth of phi A6 and phi A9 in a mutant of S. antibioticus lacking the endonuclease activity was analyzed. It is concluded that the enzyme can assume, under some circumstances, a role in in vivo restriction.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braña A. F., Wolfe S., Demain A. L. Ammonium repression of cephalosporin production by Streptomyces clavuligerus. Can J Microbiol. 1985 Aug;31(8):736–743. doi: 10.1139/m85-138. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Hopwood D. A., Kieser T., Thompson C. J. Gene cloning in Streptomyces. Curr Top Microbiol Immunol. 1982;96:69–95. doi: 10.1007/978-3-642-68315-2_5. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Wilde L. C. Restriction of a bacteriophage of Streptomyces albus G involving endonuclease SalI. J Bacteriol. 1976 Nov;128(2):644–650. doi: 10.1128/jb.128.2.644-650.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Wilde L. C. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol. 1980 Feb;116(2):323–334. doi: 10.1099/00221287-116-2-323. [DOI] [PubMed] [Google Scholar]
- Kohring G. W., Mayer F., Mayer H. Immunoelectron microscopic localization of the restriction endonuclease EcoRI in Escherichia coli BS 5. Eur J Cell Biol. 1985 May;37:1–6. [PubMed] [Google Scholar]
- Lomovskaya N. D., Chater K. F., Mkrtumian N. M. Genetics and molecular biology of Streptomyces bacteriophages. Microbiol Rev. 1980 Jun;44(2):206–229. doi: 10.1128/mr.44.2.206-229.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A., Halford S. E. The SalGI restriction endonuclease. Enzyme specificity. Biochem J. 1982 Apr 1;203(1):93–98. doi: 10.1042/bj2030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A., Halford S. E. The SalGI restriction endonuclease. Mechanism of DNA cleavage. Biochem J. 1982 Apr 1;203(1):85–92. doi: 10.1042/bj2030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A., Halford S. E. The SalGI restriction endonuclease. Purification and properties. Biochem J. 1982 Apr 1;203(1):77–84. doi: 10.1042/bj2030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A., Cohen S. N. Phenotypically cryptic EcoRI endonuclease activity specified by the ColE1 plasmid. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1265–1269. doi: 10.1073/pnas.75.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J. A., Venema G. Transformation-deficient mutants of Bacillus subtilis impaired in competence-specific nuclease activities. J Bacteriol. 1982 Oct;152(1):166–174. doi: 10.1128/jb.152.1.166-174.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós L. M., Hardisson C., Salas J. A. Isolation and properties of Streptomyces spore membranes. J Bacteriol. 1986 Mar;165(3):923–928. doi: 10.1128/jb.165.3.923-928.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1984;12 (Suppl):r167–r204. doi: 10.1093/nar/12.suppl.r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sànchez J., Barbès C., Hernandez A., de los Reyes Gavilàn C. R., Hardisson C. Restriction-modification systems in Streptomyces antibioticus. Can J Microbiol. 1985 Oct;31(10):942–946. doi: 10.1139/m85-177. [DOI] [PubMed] [Google Scholar]
- Yanagida T., Ogawara H. Deoxyribonucleases in Streptomyces. J Antibiot (Tokyo) 1980 Oct;33(10):1206–1207. doi: 10.7164/antibiotics.33.1206. [DOI] [PubMed] [Google Scholar]