Abstract
We report here the reconstitution of a pathway that leads to the apoptotic changes in nuclei by using recombinant DNA fragmentation factor (DFF), a heterodimeric protein of 40 and 45 kDa. Coexpression of DFF40 and DFF45 is required to generate recombinant DFF, which becomes activated when DFF45 is cleaved by caspase-3. The cleaved fragments of DFF45 dissociate from the DFF40, the active component of DFF. Purified DFF40 exhibited an intrinsic DNase activity that was markedly stimulated by chromatin-associated proteins histone H1 and high mobility group proteins. DFF40 also triggered chromatin condensation when incubated with nuclei. These data suggest that DFF40 is sufficient to trigger both DNA fragmentation and chromatin condensation during apoptosis.
Cells undergoing apoptosis undergo distinct morphologic changes (1). Among these changes, chromatin condensation and DNA fragmentation within the nuclei of dying cells are the most recognized markers of apoptosis (2–4). These changes in chromatin DNA are incompatible with cell survival and may mark the point of no return for execution of the cellular apoptotic pathway.
Studies in both Caenorhabditis elegans and mammalian cells have begun to reveal a conserved biochemical pathway that provides the molecular bases for the characteristic changes in cells undergoing apoptosis (5). Initiation of this apoptotic pathway leads to the activation of a group of cysteine proteases, called caspases, that cleave proteins after aspartic acid residues (6–9). Caspases usually exist in all living cells as inactive precursors that become activated when cells receive a signal to undergo apoptosis (10–13). Activated caspases cleave and disable many important cellular proteins (14). They also cleave and activate a heterodimeric protein composed of 40- and 45-kDa subunits and designated DNA fragmentation factor (DFF), which mediates the genomic DNA degradation into nucleosomal fragments (15). Both DFF40 and DFF45 are encoded by previously uncharacterized genes whose gene products do not share significant homology with other proteins of known function (15, 16).
Recently, Enari and co-workers (17, 18) showed that a mouse protein of 40 kDa, termed CAD (caspase-activated DNase), together with the mouse homologue of human DFF45, termed ICAD (inhibitor of CAD), was capable of generating a caspase-3-activated DNase activity that cleaves DNA. Mouse CAD/ICAD and human DFF40/45 therefore represent a direct link between caspase activation and DNA fragmentation.
Caspase-3 cleaves DFF45 and ICAD at two conserved cleavage sites, an event that activates DFF and CAD, respectively (15, 17). The purified DFF from HeLa cell extracts showed little DNase activity when incubated with naked DNA and caspase-3 even though the same reaction mixture induced DNA fragmentation when incubated with isolated nuclei (15). Moreover, Enari et al. (17) did detect CAD-dependent (DFF40) DNase activity when assayed in a relatively crude system. It therefore seemed likely that additional protein(s) in nuclei were required to generate a nuclease activity that cleaved chromatin DNA at the internucleosomal linker regions during apoptosis.
We report here the molecular cloning of the cDNA that encodes human DFF40 and the reconstitution of the DFF activation pathway by using purified recombinant DFF. Both DFF40 and DFF45 are required to generate active DFF. The active form of DFF, however, consists of only DFF40, with fragments of DFF45 dissociating from DFF40 after caspase-3 cleavage. The activated pure DFF exhibits a low intrinsic DNase activity when directly incubated with plasmid DNA. Such an activity was markedly stimulated when chromatin-associated proteins, such as histone H1 and high mobility group (HMG) proteins, were included in the reaction mixture. Surprisingly, activated DFF40 by itself also induces chromatin condensation when incubated with nuclei, indicating that DFF40 is the protein that triggers both DNA fragmentation and chromatin condensation during apoptosis. Activation of DFF is thus sufficient to cause the genetic death of cells undergoing apoptosis.
MATERIALS AND METHODS
Assay for DFF Activity.
Caspase-3 was expressed and purified through a nickel affinity column as described in ref. 19. DFF activity was assayed as described in ref. 15. In brief, the activity of DFF was assayed by incubating an aliquot (7 μl) of hamster nuclei (8.5 × 107 nuclei per ml) with the indicated enzyme fractions at 37°C for 2 hr in a final volume of 60 μl adjusted with buffer A (20 mM Hepes–KOH, pH 7.5/10 mM KCl/1.5 mM MgCl2/1 mM sodium EDTA/1 mM sodium EGTA/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride).
Production of DFF40 Fusion Protein.
cDNA cloning of DFF40 and production of DFF40 fusion protein were reported previously (16). In brief, degenerate oligonucleotides were designed according to the peptide sequences obtained from Edman degradation of DFF40 and used in a polymerase chain reaction (PCR) to amplify a cDNA library prepared from HeLa cells. A 250-bp PCR product was obtained and used to screen the HeLa λ Exlox cDNA library (Novagen). The longest clone (2.8 kb) that contained the entire ORF was sequenced in both strands. A 1.0-kb fragment containing the entire coding region of DFF40 cDNA was subcloned in-frame into the NdeI/XhoI sites of the bacterial expression vector pET-15b (Novagen). The expression plasmid was transformed into Escherichia coli strain BL21(DE3), and DFF40 was expressed and purified as described in ref. 16.
Western Blot Analysis.
A monoclonal antibody against FLAG tag was from Kodak Scientific Imaging System. Anti-DFF40 antiserum was generated by immunizing rabbits with a recombinant DFF40 fusion protein generated as described above. Immunoblot analysis was performed with the horseradish peroxidase-conjugated goat anti-mouse (FLAG tag) or goat anti-rabbit (DFF45) IgG by using enhanced chemiluminescence (ECL) Western blotting detection reagents (Amersham).
Expression and Purification of Recombinant DFF.
Expression of DFF45 and DFF40 in one plasmid was done in principle as described by Khokhlatchev et al. (20). Two primers were used to PCR amplify the coding region of DFF40 cDNA. The resulting 1-kb DNA fragment was subcloned into the XbaI and XhoI sites of the bacterial expression vector pET-15b (Novagen), and the resulting plasmid was designated pET-15b-DFF40His. Two primers were then used to PCR-amplify the coding region of DFF45, and the resulting DNA fragment was digested with XhoI and ligated into pET-15b-DFF40His digested with XhoI. The expression plasmid containing both DFF45 and DFF40 was used to transform E. coli BL21(pLys) (Novagen). The expression of DFF was induced by isopropyl β-d-thiogalactoside (IPTG), and the recombinant DFF was purified through a nickel affinity column followed by a Mono S 5/5 column (Pharmacia). About 0.2 mg of DFF was purified from a 500-ml culture.
Expression of DFF in 293 Cells.
A FLAG tag was engineered at the carboxyl terminus of DFF40 by PCR amplifying the 2.8-kb cDNA containing the entire coding region of DFF40. The PCR fragment was subcloned into the EcoRV and XhoI sites of a pcDNA3 vector (Invitrogen) digested with the same restriction enzymes. The resulting plasmid was designated pDFF40-flag. The 1.5-kb cDNA encoding the entire coding region of DFF45 was subcloned into the EcoRI sites of a pcDNA3 vector (Invitrogen), and the resulting plasmid was designated pDFF45. Human embryonic kidney 293 cells were set up and transfected with 20 μg of vector alone, 10 μg of pDFF40-flag plus 10 μg of vector, 10 μg of pDFF45 plus 10 μg of vector, or 10 μg of pDFF40-flag plus 10 μg of pDFF45 by using an MBS transfection kit (Stratagene) as described (21). After 24 hr, the cells were harvested and the S-100 fractions were prepared as described in ref. 19. The assay for DFF was performed as described in ref. 15.
Immunostaining.
Simian CV-1 cells were transfected with 10 μg of pDFF45 plus 10 μg of pcDNA3 vector, pDFF40-flag plus 10 μg of pcDNA3 vector, or 10 μg of pDFF45 plus 10 μg of DFF40-flag by the calcium phosphate precipitation method. Twenty-four hours after transfection, CV-1 cells were immunostained with anti-FLAG antibody or anti-DFF45 antibody as described in ref. 22. The antigen-antibody complexes were probed with fluorescein 5-isothiocyanate (FITC)-conjugated goat anti-rabbit (for DFF45) IgG (1:1000) or goat anti-mouse (for DFF40) IgG (1:1000). The cells were washed three times with PBS before being examined under a fluorescence microscope.
RESULTS
Expression of Recombinant DFF Activity and Immunolocalization of DFF45 and DFF40.
In previous studies we purified DFF from HeLa cell extracts as a heterodimer of 40- and 45-kDa subunits. We cloned and completely sequenced the 45-kDa subunit (15). Four peptides from the 40-kDa subunit were also sequenced, including the amino terminus of the protein (Fig. 1A). On the basis of the sequences of these peptides, we obtained a cDNA clone encoding the entire DFF40 protein by screening a cDNA library (16).
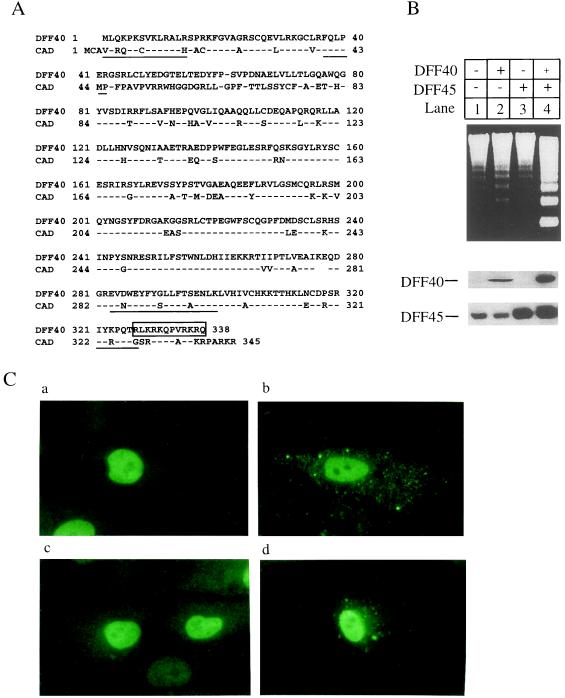
Figure 1.
Expression of DFF and nuclear localization of DFF40 and DFF45. (A) Amino acid sequences of human DFF40 and mouse CAD (16, 17) are aligned by the Lipman–Pearson method of the dnastar program. The proteolytic peptide sequences from purified DFF40 are underlined. A putative nuclear localization signal at the carboxyl terminus of DFF40 is boxed. The amino acid residues are numbered on the left and right. (B) Human embryonic kidney 293 cells were grown and transfected as described in ref. 21. The 293 cells were transfected with the vector alone (lanes 1), pDFF40-flag alone (lanes 2), pDFF45 alone (lanes 3), or pDFF40-flag plus pDFF45 (lanes 4). The cells were harvested 36 hr after transfection and the S-100 fraction was prepared as described in ref. 19. Aliquots (80 μg) of cytosolic proteins were incubated in the presence of caspase-3 with aliquots (7 μl) of hamster liver nuclei for 2 hr at 37°C in 60 μl of buffer A. Genomic DNA was isolated and analyzed as described in ref. 15. Aliquots (80 μg) of S-100 prepared as described above were also subjected to SDS/10% PAGE and transferred to nitrocellulose filters. The filters were probed either with a rabbit anti-DFF45 antiserum (15) or with M2 anti-FLAG antiserum (1:2000). The antigen–antibody complexes were visualized with an ECL method. (C) CV-1 cells were transfected with pDFF40-flag alone (b), pDFF45 alone (a), or pDFF40-flag plus pDFF45 (c and d). The cells were fixed and subsequently probed with the rabbit anti-DFF45 antiserum or the M2 anti-FLAG antibody. The antigen–antibody complexes were visualized by a fluorescein isothiocyanate method. (a) Cells were transfected with pDFF45 and stained with anti-DFF45 antibody. (b) Cells were transfected with pDFF40-flag and stained with anti-FLAG antibody. (c) Cells were transfected with pDFF45 plus pDFF40-flag and stained with anti-DFF45 antibody. (d) Cells were transfected with pDFF45 plus pDFF40-flag and stained with anti-FLAG antibody.
Fig. 1A shows the sequence of human DFF40 protein in comparison with that of mouse CAD. The two proteins are about 71% identical throughout the entire sequence, with a noticeable exception of the region between amino acids 41 and 79, which differs completely between the two proteins. This difference is the result of a change in the reading frame, which could be because of a sequencing error in the reported CAD cDNA (17). The putative nuclear localization signal previously noticed in the carboxyl terminus of CAD is conserved in DFF40 (17). To test the ability of recombinant DFF to induce DNA fragmentation, we expressed the cDNAs encoding DFF45 and DFF40 either individually or together by transfection into human embryonic kidney 293 cells. Extracts from the transfected cells were incubated with hamster liver nuclei in the presence of active caspase-3. Neither DFF40 nor DFF45 expressed alone exhibited a significant increase in DNA fragmentation activity when incubated with nuclei compared with a vector control (Fig. 1B, lanes 1–3), even though both proteins were expressed as measured by Western blot analysis (Fig. 1B). The antibody against DFF45 also recognized the endogenous protein. However, when the two proteins were coexpressed, a dramatic increase in DFF activity was observed (Fig. 1B, lane 4). The activity of recombinant DFF was dependent on caspase-3, because no activity was observed when caspase-3 was omitted from the reaction (data not shown).
DFF activity was previously detected and purified in the 100,000 × g supernatant (S-100), which suggested that latent DFF might be sequestered in the cytosol (15). On the basis of similar observations, Enari et al. (17) proposed that the activation of DNA fragmentation is a result of releasing the cytosolic retention of CAD (DFF40) by ICAD (DFF45) similar to the activation of NF-κB. Unexpectedly, as shown in Fig. 1C, when measured by immunofluorescence staining, the majority of the staining for both DFF40 and DFF45 was observed in nuclei when expressed by transient transfection either alone or together in CV-1 cells. Such nuclear localization of DFF has been confirmed in several other cell lines, including HeLa cells, simian virus 40-transformed human fibroblast cells, and human embryonic kidney 293 cells (data not shown).
Delineation of the DFF Activation Pathway.
Latent DFF containing both 40- and 45-kDa subunits becomes activated when DFF45 is cleaved by caspase-3 (15). Because coexpression of DFF40 and DFF45 is required to generate functional recombinant DFF (Fig. 1B), we engineered an E. coli expression plasmid that contains both DFF40 and DFF45 cDNAs with DFF40 tagged with six histidine residues at its carboxyl terminus. This double-expression system generated functional DFF, which could be purified through a nickel affinity column. As shown in Fig. 2A, we were able to generate recombinant DFF by using this expression system, and we purified the recombinant DFF to homogeneity (Fig. 2C, lane 1). Recombinant DFF induced DNA fragmentation when incubated with nuclei in the presence of caspase-3, and an inhibitor of caspase-3 blocked this activity (Fig. 2A).
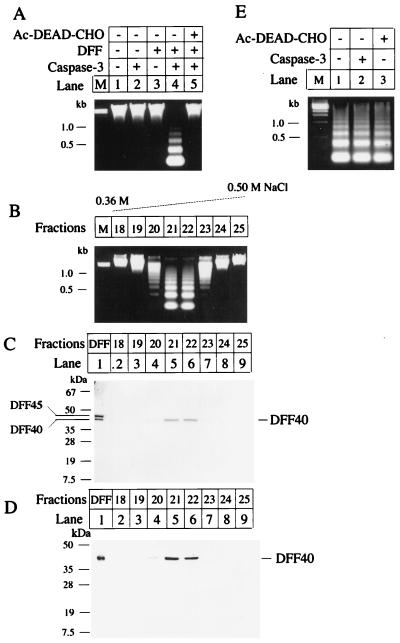
Figure 2.
DFF activation pathway. (A) Recombinant DFF was expressed and purified. An aliquot (7 μl) of hamster liver nuclei was incubated alone (lane 1), with 50 ng of caspase-3 (lane 2), with 500 ng of purified DFF (lane 3), or with 50 ng of caspase-3 plus 500 ng of purified DFF (lane 4) at 37°C for 2 hr in a final volume of 60 μl of buffer A. In lane 5, 50 ng of caspase-3 was preincubated with 1 μM Ac-DEAD-CHO (a tetrapeptide aldehyde inhibitor of caspase-3) at 37°C for 5 min followed by the addition of 500 ng of DFF and 7 μl of nuclei and incubation for 2 hr at 37°C in 60 μl of buffer A. The genomic DNAs were isolated and analyzed as described in ref. 15. (B) Recombinant DFF was generated and purified as described. An aliquot of 1 mg was incubated with 100 μg of caspase-3 at 30°C for 1 hr. The sample was then purified through a Mono Q 5/5 column (Pharmacia) followed by a Mono S 5/5 column (Pharmacia). The column was eluted with a 30 ml 0.3–0.5 M linear NaCl gradient. Fractions of 1 ml were collected. An aliquot (20 μl) of each fraction was incubated with an aliquot of 7 μl of hamster liver nuclei at 37°C for 2 hr in 60 μl of buffer A. The genomic DNA was isolated, analyzed, and visualized as described in ref. 15. (C) An aliquot (1 μg) of purified recombinant DFF (lane 1) and aliquots of 30 μl of the indicated Mono S column fractions were subjected to SDS/12% PAGE followed by Coomassie brilliant blue staining. The positions of DFF40 and DFF45 are indicated. (D) An aliquot (20 μl) of the indicated Mono S column fractions was subjected to SDS/12% PAGE and transferred to a nitrocellulose filter. The filter was probed with a rabbit anti-DFF40 antiserum (1:2000) and the antigen–antibody complexes were visualized through an ECL method. (E) An aliquot (30 μl) of the active DFF purified through the Mono S column was incubated with 7 μl of hamster liver nuclei at 37°C for 2 hr in 60 μl of buffer A with buffer alone (lane 1) or in the presence of 50 ng of caspase-3 (lane 2) or 10 μM Ac-DEAD-CHO (lane 3). The genomic DNA was isolated, analyzed, and visualized as described in ref. 15.
To identify the active component of DFF, we reproduced the DFF activation process in vitro by incubating the pure recombinant DFF with caspase-3 and purified the active component of DFF by subjecting the reaction mixture to Mono Q column chromatography followed by Mono S column chromatography. The fractions eluted from these two columns were assayed for DFF activity by incubating them with hamster liver nuclei without further addition of caspase-3. As shown in Fig. 2 B and C, the fractions that exhibited constitutive DFF activity consisted of only the 40-kDa subunit, indicating that DFF40 is the active form of DFF. The identity of the active DFF as DFF40 was confirmed by Western blot analysis using an antibody generated against DFF40 recombinant protein (Fig. 2D). The fragments of DFF45 generated by caspase-3 cleavage were observed in the column fractions that did not associate with DFF activity (data not shown). Active DFF purified through the Mono S column induced DNA fragmentation in coincubated nuclei without a further requirement for caspase-3 (Fig. 2E). The activity was insensitive to the caspase-3 inhibitor (Fig. 2E). Hereafter, we designate the activated pure DFF40 as A-DFF40.
Stimulation of DNase Activity of DFF40 by Chromatin-Associated Proteins.
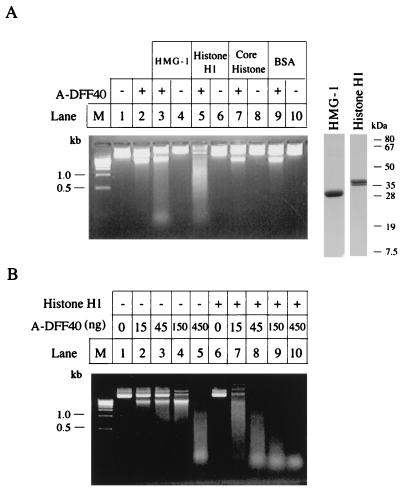
Purified DFF from HeLa cell extracts showed little nuclease activity in the presence of caspase-3 when incubated directly with naked DNA compared with incubation with nuclei (15). However, Enari et al. (17) did detect CAD-dependent (DFF40) DNase activity when assayed in a relatively crude system. It seemed likely that additional protein(s) were required to generate a nuclease activity that cleaved chromatin DNA at the internucleosomal linker regions during apoptosis. We therefore set up an assay to search for additional protein factors from HeLa cell high-salt nuclear extracts that may work together with activated DFF to generate high DNase activity. We identified and purified such a protein from HeLa cell nuclear extracts, and protein sequencing analysis revealed that it was HMG protein 2 (P.L. and X.W., unpublished data). That prompted us to test other chromatin-associated proteins, such as core histones, histone H1, and other HMG proteins for stimulation of DFF-dependent DNase activity. As shown in Fig. 3A, histone H1 and HMG-1 stimulated the DNase activity of DFF40 when incubated with plasmid DNA, whereas the core histones and bovine serum albumin (BSA) showed little effect. To demonstrate further the stimulatory effects of chromatin-associated proteins, we assayed the DNase activity of DFF40 with increasing concentrations of A-DFF40 in the presence and absence of histone H1. As shown in Fig. 3B, A-DFF40 manifested its intrinsic DNase activity only when high concentrations of it were used (Fig. 3B, lane 5). Little DNase activity was detected at lower concentrations unless histone H1 was present (Fig. 3B, lanes 2–4 and 7–10). The stimulatory effect of histone H1 was more than 10-fold.
Figure 3.
Stimulation of DNase of DFF40 by histone H1 and HMG-1. (A) An aliquot (2 μg) of plasmid DNA (pcDNA3 vector) was incubated with either a 45-ng aliquot of A-DFF40 purified through the Mono S column as described in Fig. 2 in 30 μl of buffer A at 37°C for 2 hr alone (lane 2) or in the presence of 1 μg of bovine HMG-1 (lane 3), 0.5 μg of bovine histone H1 (lane 5), 1 μg of core histone (lane 7), or 1 μg of BSA (lane 9). The plasmid DNA was also incubated with buffer alone (lane 1), 1 μg of bovine HMG-1 (lane 4), 0.5 μg of histone H1 (lane 6), 1 μg of core histone, or 1 μg of BSA (lane 10). The reactions were stopped by adding 5 mM EDTA and the products were directly loading onto a 1.2% agarose gel containing 5 μg/ml ethidium bromide. An aliquot (5 μg) of bovine HMG-1 or histone H1 was also directly subjected to SDS/PAGE followed by Coomassie brilliant blue staining (the right two lanes with molecular mass markers). (B) An aliquot (2 μg) of plasmid DNA was incubated with the indicated amount of A-DFF40 purified through the Mono S column as described in the legend Fig. 2 in the absence (lanes 2–5), or presence (lanes 6–10) of 0.5 μg of histone H1 in a final volume of 30 μl of buffer A at 37°C for 2 hr. The reactions were stopped by adding 5 mM EDTA and the products were directly loaded onto a 1.2% agarose gel containing 5 μg/ml ethidium bromide.
Activated DFF40 Triggers Chromatin Condensation.
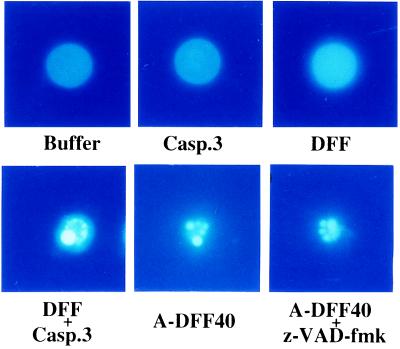
Chromatin condensation is one of the morphological hallmarks of apoptosis (1–4). The molecular mechanism for apoptosis-associated chromatin condensation, however, is unknown. Unexpectedly, when latent DFF plus caspase-3, or A-DFF40, was incubated with isolated nuclei, chromatin DNA became condensed into several small bright granules, a typical nuclear morphology observed when cells are undergoing apoptosis (ref. 2; Fig. 4). The control nuclei, on the other hand, demonstrated uniform DNA staining throughout the nuclear interior (Fig. 4). Addition of caspase-3 or latent DFF alone had no effect on the DNA staining patterns of these nuclei. A general caspase inhibitor, the modified tripeptide z-VAD-fmk, had no effect on DNA condensation when A-DFF40 was used, indicating that DFF is different from AIF, a z-VAD-sensitive mitochondrial protein that triggers apoptosis in nuclei (23).
Figure 4.
A-DFF40 induces chromatin condensation in nuclei. An aliquot (7 μl) of hamster liver nuclei was incubated at 37°C for 2 hr with buffer alone, with 50 ng of caspase-3, with 500 ng of DFF, with 50 ng of caspase-3 plus 500 ng of DFF, with 300 ng of A-DFF40 purified through the Mono S column as described in the legend of Fig. 2, or with 300 ng of A-DFF40 plus 20 μM z-VAD-fmk as indicated. The nuclei were then stained with 4′,6-diamidino-2-phenylindole (DAPI) and observed under a fluorescence microscope with a DAPI filter.
DISCUSSION
We demonstrated above that DFF is cleaved and activated by caspase-3 at two sites that were mapped previously (15). After cleavage, the cleaved fragments of the DFF45 subunit are no longer associated with the DFF40 subunit (A-DFF40), which is the active component of DFF. A-DFF40 is sufficient to induce chromatin DNA fragmentation and condensation when incubated with nuclei without further requirement for caspase activity. Such an activation pathway is consistent with the model proposed by Enari et al. (17) that CAD (mouse homologue of DFF40) is the active component that induces DNA fragmentation.
High levels of DFF40 and DFF45 are detected in normal nuclei by immunofluorescence staining (Fig. 1C). The observation that DFF40 and DFF45 are present in nuclei of nonapoptotic cells suggests that the cytosolic retention model for activation of DFF is not correct. The exclusive presence of DFF in the cytosolic fraction after biochemical fraction is the result of leakage from nuclei during homogenization. Indeed, biochemical fractionation of these transfected cells revealed that DFF40 and DFF45 exist almost exclusively in the cytosolic fraction (data not shown). Such nuclear leakage during homogenization has also been previously observed for other nuclear proteins (24). DFF45 is therefore like other nuclear substrates of caspase-3 such as poly(ADP-ribose) polymerase and lamins that are cleaved as the result of nuclear translocation of active caspases (25, 26).
Without coexpression of DFF45, DFF40 produced in either mammalian cells or insect cells does not have DFF activity (Fig. 2; X.L and X.W., unpublished observation). DFF45 therefore seems to mediate the correct folding of DFF40 but is not the component of the activated DFF. Such a function by definition is that of a molecular chaperone (27). However, unlike the general molecular chaperones such as Hsp70 and Hsp60 systems (27, 28), DFF45 remains complexed with DFF40, keeping it from being active until a specific signal is received, in this case, the activation of caspase-3. Such a specific molecular chaperone provides a double safety control to prevent unwanted activation of DFF. First, such an arrangement prevents newly synthesized DFF40 from cleaving DNA. Because DFF40 is translocated to nuclei (Fig. 3C), imbalanced expression of DFF45 and DFF40 could otherwise have a catastrophic effect on the cells by introducing chromatin condensation and degradation of DNA. Only the DFF40 that is complexed with DFF45 has the potential to generate active DFF. Second, by forming a complex with DFF40, DFF45 also prevents DFF40 from activating its DNase activity. In this way, only the DFF heterodimer that is cleaved by caspase-3 will become active. In this sense, DFF45 is also an inhibitor for DFF40, as proposed for ICAD (17, 18). However, unlike the result with ICAD, we found that overexpression of wild-type DFF45 in stably transfected CHO cells did not inhibit their DFF activity (X.L. and X.W., unpublished observation).
A-DFF40 has an intrinsic DNase activity when incubated with plasmid DNA (Fig. 3B), which confirms the proposal of Enari et al. (17) that the mouse homologue of DFF40 (CAD) is the caspase-activated DNase. However, the reason we previously failed to see this activity when we used purified DFF from HeLa cells is that DFF is a poor DNase when directly incubated with naked DNA, so that a much higher concentration of DFF was needed to cleave naked DNA than chromatin DNA in nuclei (ref. 15; Fig. 3). This observation suggested that additional nuclear proteins contribute to DNA fragmentation. Indeed, we found that abundant chromatin-associated proteins, such as HMG-1, HMG-2, and histone H1, can markedly stimulate the intrinsic DNase activity of DFF40 (Fig. 3; P.L. and X.W., unpublished observation). This observation suggests that rather than passively cleaving chromatin DNA at the linker region because it is more accessible to the nuclease, DFF-dependent DNase is targeted directly to the nucleosomal linker region of chromatin, where the HMG proteins and histone H1 are known to be located (29–31). In addition, because histone H1 and HMGs are also believed to be involved in organizing higher-order chromatin structure (29), these proteins may also facilitate A-DFF40 DNase to disassemble these higher-order chromatin structures. Such a model would provide a more efficient way to disassemble complex chromatin DNA into nucleosomes compared with disassembly by random cleavage of DNA. The molecular mechanism by which these chromatin-associated proteins stimulate A-DFF40 DNase activity should be an interesting topic for future study.
Surprisingly, when A-DFF40 was added to nuclei, their chromatin also become condensed, another hallmark feature of apoptosis (4). This result puts DFF in a central position responsible for the observed nuclear changes during apoptosis. It is possible that DFF triggers chromatin condensation by cleaving chromatin DNA. However, it has been reported that treatment of isolated nuclei with micrococcal nuclease induces nucleosomal DNA fragmentation but nevertheless has no effect on chromatin condensation (32). It is interesting to note that cleavage of DNA at the nucleosomal linker region by micrococcal nuclease is blocked, not facilitated, by the presence of histone H1 (33). It is also possible that A-DFF40 activates an independent pathway leading to chromatin condensation. All these become testable hypotheses now with the availability of pure recombinant DFF.
Acknowledgments
We thank Ms. Yucheng Li for excellent technical assistance. We thank Dr. Clive Slaughter, Ms. Carolyn Moomaw, and Mr. Steve Afendis for help with the protein sequencing analysis. We also thank Dr. R. C. Johnson (Univ. of California, Los Angeles) for providing us with purified bovine HMG proteins. We are grateful to our colleagues Drs. Joseph Goldstein, Michael Brown, Steven McKnight, and Holt Oliver for critically reading the manuscript. X.W. is also supported by grants from the American Cancer Society (RE258) and the National Institutes of Health (GMRO1–55942). W.T.G. is supported by grants from the National Institutes of Health (GMRO1–29935) and the Robert Welch Foundation (I-823).
ABBREVIATIONS
- DFF
DNA fragmentation factor
- DFF40
the 40-kDa subunit of DFF
- CAD
caspase-activated DNase
- ICAD
inhibitor of CAD
- HMG
high mobility group protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF064019).
References
- 1.Kerr J F R, Wyllie A H, Currie A R. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyllie A H. Nature (London) 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 3.Wyllie A H, Kerr J F, Currie A R. Int Rev Cytol. 1980;68:251–305. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 4.Wyllie A H, Morris R G, Smith A L, Dunlop D. J Pathol. 1984;142:66–77. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- 5.Vaux D L. Cell. 1997;90:389–390. doi: 10.1016/s0092-8674(00)80497-3. [DOI] [PubMed] [Google Scholar]
- 6.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 7.Faleiro L, Kobayashi R, Fearnhead H, Lazebnik Y. EMBO J. 1997;16:2271–2281. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins L M, Kottke T, Mesner P W, Basi G S, Sinha S, Frigon N, Tatar E, Tung J S, Bryant K, Takahashi A, Svingen P A, Madden B J, McCormick D J, Earnshaw W C, Kaufmann S H. J Biol Chem. 1997;272:7421–7430. doi: 10.1074/jbc.272.11.7421. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi A, Hirata H, Yonehara S, Imai Y, Lee K K, Moyer R W, Turner P C, Mesner P W, Okazaki T, Sawai H, Kishi S, Yamamoto K, Okuma M, Sasada M. Oncogene. 1997;14:2741–2752. doi: 10.1038/sj.onc.1201131. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson W D, Ali A, Thornberry N A, Vailancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raji S M, Smulson M E, Yamin T T, Yu V L, Miller D K. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 11.Tewari M, Quan L, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 12.Schlegel J, Peters I, Orrenius S, Miller D K, Thornberry N A, Yamin T-T, Nicholson W D. J Biol Chem. 1996;271:1841–1844. doi: 10.1074/jbc.271.4.1841. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Zelenski N G, Yang J, Sakai J, Brown M S, Goldstein J L. EMBO J. 1996;15:1012–1020. [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholson W D, Thornberry N A. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Zou H, Slaughter C, Wang X. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 16.Liu X. Ph.D. Dissertation. Atlanta, GA: Emory Univ.; 1997. pp. 135–138. [Google Scholar]
- 17.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. Nature (London) 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 18.Sakahira H, Enari M, Nagata S. Nature (London) 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 20.Khokhlatchev A, Xu S, English J, Wu P, Schaefer E, Cobb M H. J Biol Chem. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- 21.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 22.Luo X, Sawadogo M. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamzami N, Susin S A, Marchetti P, Hirsch T, Gomez-Monterrey I, Castedo M, Kroemer G. J Exp Med. 1996;183:1523–1544. doi: 10.1084/jem.183.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paine P L, Austerberry C F, Desjarlais L J, Horvitz S B. J Cell Biol. 1983;97:1240–1242. doi: 10.1083/jcb.97.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann S H, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 26.Lazebnik Y, Takahashi A, Moir R, Goldman R, Poirier G G, Kaufman S H, Earnshaw W C. Proc Natl Acad Sci USA. 1995;92:9042–9046. doi: 10.1073/pnas.92.20.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartl F U, Hlodan R, Langer T. Trends Biochem Sci. 1994;19:20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 28.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 29.Jackson J B, Pollock J M, Rill R L. Biochemistry. 1979;18:3739–3748. doi: 10.1021/bi00584a015. [DOI] [PubMed] [Google Scholar]
- 30.Peters E, Levy-Wilson B, Dixon G H. J Biol Chem. 1979;254:3358–3361. [PubMed] [Google Scholar]
- 31.Schroter H, Bode J. Eur J Biochem. 1982;127:429–436. doi: 10.1111/j.1432-1033.1982.tb06890.x. [DOI] [PubMed] [Google Scholar]
- 32.Sun D Y, Jiang S, Zheng L M, Ojcious D M, Young J D. J Exp Med. 1994;179:559–568. doi: 10.1084/jem.179.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roche J, Girardet J L, Gorka C, Lawrence J J. Nucleic Acids Res. 1985;13:2843–2853. doi: 10.1093/nar/13.8.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]