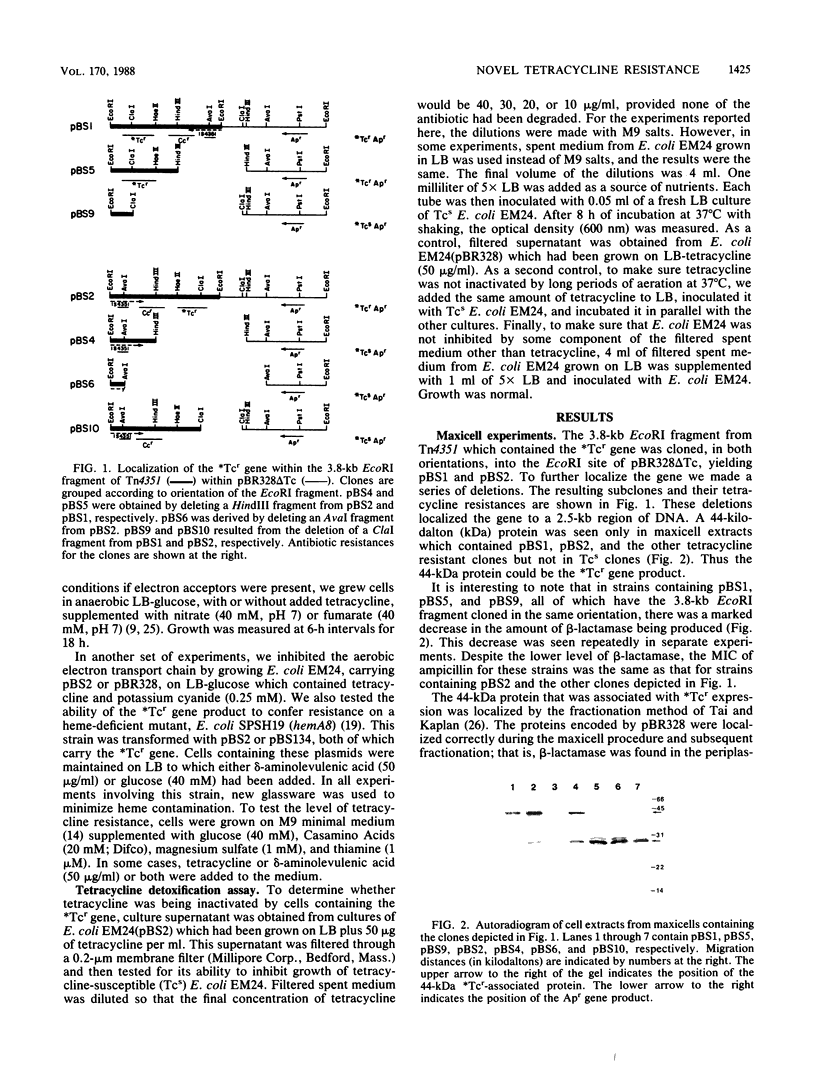
Abstract
A tetracycline resistance (Tcr) gene that was found originally on two Bacteroides plasmids (pBF4 and pCP1) confers tetracycline resistance on Escherichia coli, but only when it is grown aerobically. Using maxicells, we have identified a 44-kilodalton protein which is encoded by the region that carries the Tcr gene and which may be the Tcr gene product. Localization experiments indicate that this 44-kilodalton protein is cytoplasmic. To determine whether the tetracycline resistance gene is expressed under anaerobic conditions, we have constructed a protein fusion between the Tcr gene and lacZ. In strains of E. coli carrying the fusion, beta-galactosidase activity was the same when the cells were grown under anaerobic conditions as when the cells were grown under aerobic conditions. This indicates that the tetracycline resistance gene product is made under anaerobic conditions but does not work. The failure of the Tcr protein to function under anaerobic conditions was not due to a requirement for function of the anaerobic electron transport system, because neither nitrate nor fumarate added to anaerobic media restored tetracycline resistance. Inhibition of the aerobic electron transport system with potassium cyanide did not prevent growth on tetracycline of cells containing the Tcr gene. A heme-deficient mutant, E. coli SHSP19, which carries the Tcr gene, was still resistant to tetracycline even when grown in heme-free medium. These results indicate that functioning of the Tcr gene product is not dependent on the aerobic electron transport system. Thus the requirement for aerobic conditions appears to reflect a requirement for oxygen. Spent medium from an E. coli strain carrying the Tcr gene, which was grown in medium containing tetracycline (50 micrograms/ml), did not inhibit growth of a tetracycline-susceptible strain of E. coli. Thus, the Tcr gene product may be detoxifying tetracycline.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman M. L., Jackson D. E. Selection of lac gene fusions in vivo: ompR-lacZ fusions that define a functional domain of the ompR gene product. J Bacteriol. 1984 Aug;159(2):750–756. doi: 10.1128/jb.159.2.750-756.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J Bacteriol. 1986 Feb;165(2):564–569. doi: 10.1128/jb.165.2.564-569.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Howe T. G. Bacterial resistance to the tetracyclines. Microbiol Rev. 1978 Dec;42(4):707–724. doi: 10.1128/mr.42.4.707-724.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias L., Cervantes L., Covarrubias A., Soberón X., Vichido I., Blanco A., Kupersztoch-Portnoy Y. M., Bolivar F. Construction and characterization of new cloning vehicles. V. Mobilization and coding properties of pBR322 and several deletion derivatives including pBR327 and pBR328. Gene. 1981 Jan-Feb;13(1):25–35. doi: 10.1016/0378-1119(81)90040-8. [DOI] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Homology between clindamycin resistance plasmids in Bacteroides. Plasmid. 1984 May;11(3):268–271. doi: 10.1016/0147-619x(84)90035-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto F., Horigome T., Kanbayashi M., Yoshida K., Sugano H. An improved method for separation of low-molecular-weight polypeptides by electrophoresis in sodium dodecyl sulfate-polyacrylamide gel. Anal Biochem. 1983 Feb 15;129(1):192–199. doi: 10.1016/0003-2697(83)90068-4. [DOI] [PubMed] [Google Scholar]
- Jones H. M., Gunsalus R. P. Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate. J Bacteriol. 1985 Dec;164(3):1100–1109. doi: 10.1128/jb.164.3.1100-1109.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon B. R., Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987 Mar;51(1):88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. G., Guiney D. G. Characterization and mapping of regions encoding clindamycin resistance, tetracycline resistance, and a replication function on the Bacteroides R plasmid pCP1. J Bacteriol. 1986 Aug;167(2):517–521. doi: 10.1128/jb.167.2.517-521.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robillard N. J., Tally F. P., Malamy M. H. Tn4400, a compound transposon isolated from Bacteroides fragilis, functions in Escherichia coli. J Bacteriol. 1985 Dec;164(3):1248–1255. doi: 10.1128/jb.164.3.1248-1255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt R. Analysis of melibiose mutants deficient in alpha-galactosidase and thiomethylgalactoside permease II in Escherichia coli K-12. J Bacteriol. 1968 Aug;96(2):462–471. doi: 10.1128/jb.96.2.462-471.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Getty C., Gardner J. F., Salyers A. A. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986 Mar;165(3):929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Getty C., Guthrie E. P., Salyers A. A. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J Bacteriol. 1986 Jun;166(3):959–965. doi: 10.1128/jb.166.3.959-965.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Guthrie E. P., Salyers A. A., Gardner J. F. Evidence that the clindamycin-erythromycin resistance gene of Bacteroides plasmid pBF4 is on a transposable element. J Bacteriol. 1985 May;162(2):626–632. doi: 10.1128/jb.162.2.626-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Gonda M. A. Comparison of the transposon-like structures encoding clindamycin resistance in Bacteroides R-plasmids. Plasmid. 1985 May;13(3):182–192. doi: 10.1016/0147-619x(85)90041-1. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Guest J. R. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973 May;114(2):563–570. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Săsărman A., Surdeanu M., Horodniceanu T. Locus determining the synthesis of delta-aminolevulinic acid in Escherichia coli K-12. J Bacteriol. 1968 Nov;96(5):1882–1884. doi: 10.1128/jb.96.5.1882-1884.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tally F. P., Snydman D. R., Shimell M. J., Malamy M. H. Characterization of pBFTM10, a clindamycin-erythromycin resistance transfer factor from Bacteroides fragilis. J Bacteriol. 1982 Aug;151(2):686–691. doi: 10.1128/jb.151.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock G. M., Berman M. L., Silhavy T. J. Chimeric genetics with beta-galactosidase. Gene Amplif Anal. 1983;3:27–64. [PubMed] [Google Scholar]
- Welch R. A., Macrina F. L. Physical characterization of Bacteroides fragilis R plasmid pBF4. J Bacteriol. 1981 Feb;145(2):867–872. doi: 10.1128/jb.145.2.867-872.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]




