Abstract
The amino-terminal region of the low-density lipoprotein receptor (LDLR) contains seven imperfect repeats of a cysteine-rich, roughly 40-aa module (LDL-A module) that are critical for apolipoprotein binding. LDL-A modules are found in numerous cell-surface and secreted proteins and are believed to mediate extracellular protein–protein interactions. The cellular receptor for subgroup A Rous sarcoma virus (RSV) contains a single LDL-A module that binds the RSV envelope protein and allows viral infection. To define residues in an LDL-A module responsible for ligand recognition, we used a gain of function assay by using a chimeric protein in which the LDL-A module of Tva was replaced with a highly homologous module from human LDLR (LDL-A4) and determined whether this chimera or mutants produced in it could mediate RSV infection. LDL-A4 does not function as an RSV receptor; however, systematic replacement of the nonconserved residues of the LDL-A4 module in the chimeric protein with the corresponding residues from Tva identified three residues sufficient to alter ligand specificity and convert LDL-A4 to an efficient viral receptor. Mutations of the corresponding residues in the Tva LDL-A module decreased both envelope binding and viral receptor function, confirming the importance of these residues in ligand recognition by this module. Analysis of the hLDL-A5 structure demonstrates that these three residues are clustered at one end of the LDL-A module. These results demonstrate that using a single LDL-A module in a gain of function assay is a useful model to investigate ligand recognition by this module.
The human low-density lipoprotein receptor (hLDLR) is a cell-surface glycoprotein that regulates plasma cholesterol levels by mediating the uptake of low-density lipoprotein, the major cholesterol transport protein in human plasma. The ligand-binding domain of hLDLR is determined by seven imperfect repeats in the extracellular region of the protein, each approximately 40 aa long and containing three disulfide bonds (LDL-A modules; refs. 1 and 2). Each module is thought to fold independently, and different modules or combinations of modules interact with various ligands to determine receptor specificity (2). The carboxyl terminus of each LDL-A module contains a cluster of negatively charged amino acids required for receptor function and postulated to bind to positively charged residues in lipoprotein at least in part via ionic interactions (1, 2). Analysis of the molecular requirements for LDLR module function has relied on the numerous familial hypercholesterolemia mutations known to affect LDLR function (3). However, a large number of these mutations affect presentation of LDLR on the cell surface as well as ligand binding, thereby complicating analysis of contributions of specific residues to ligand binding.
LDL-A modules are found in numerous cell-surface and secreted proteins and are believed to mediate extracellular protein–protein interactions (4). The cellular receptor for subgroup A Rous sarcoma virus (RSV; an avian retrovirus) is a small membrane-associated protein, Tva, containing a single LDL-A module that binds the RSV envelope protein and allows viral infection (5–10). The 40-aa module from Tva is sufficient for full receptor activity (5, 8), indicating that this region of Tva determines the specificity of ligand (RSV-A envelope) binding. Mutational analysis of the acidic residues near the carboxyl terminus of the Tva LDL-A module demonstrated that they are important for envelope binding (8, 9). Several basic residues in RSV-A envelope (EnvA) are required for efficient binding to Tva, suggesting that the Tva-EnvA interaction may be mechanistically similar to the binding of LDLR to its ligands, having as at least one determinant the ionic interaction of acidic residues in the receptor and the basic residues on the virus (11). Because receptor specificity and function of Tva are determined by a single LDL-A module, we used Tva as a model system to investigate the determinants of ligand specificity for the LDL-A module.
In this study, we defined residues in the Tva LDL-A module involved in ligand recognition by substituting a closely related sequence from the human LDLR (module A4) for the Tva module and then converting the hLDL-A4 module into an RSV-A receptor by alteration of individual residues. Our results demonstrate that substitution of three residues from Tva for the corresponding regions in hLDL-A4 renders the chimeric receptor fully functional as an RSV-A receptor. Furthermore, mutation of wild-type (wt) Tva at the corresponding residues impaired ligand binding. Thus, by using a gain of function assay we have identified residues in an LDL-A module that determine ligand specificity and viral receptor function.
MATERIALS AND METHODS
Cells and Viruses.
Human embryonic kidney 293T cells and quail cell line QT6 were maintained as described previously (5, 9, 10). HeLa cells were maintained as suspension cultures in Ham’s F-12 media (GIBCO) containing 10% (vol/vol) bovine calf serum. RCAS(A)AP viral stocks and vaccinia virus stocks were generated as described previously (5, 12).
DNA Methodology.
To facilitate identification of Tva/LDL-A4 chimeric proteins, Tva was epitope-tagged at the N terminus with two tandem copies of myc-tag, which can be recognized by an mAb, 9E10. A Tva expression plasmid containing a unique BamHI site upstream of the LDL-A motif was modified by inserting into the BamHI site of oligonucleotides OS242 (5′-GATCAGAACAAAAGCTTATTTCTGAAGAAGATCTTG-3′) and OS243 (5′-GATCCAAGATCTTCTTCAGAAATAAGCTTTTGTTCT-3′) encoding the myc epitope tag. The resulting plasmid Myc-Tva has an insertion of 22 residues, EQKLISEEDLGSEQKLISEEDL, where the myc-Tag sequences are underlined, upstream of the LDL-A module of Tva (Fig. 1B).
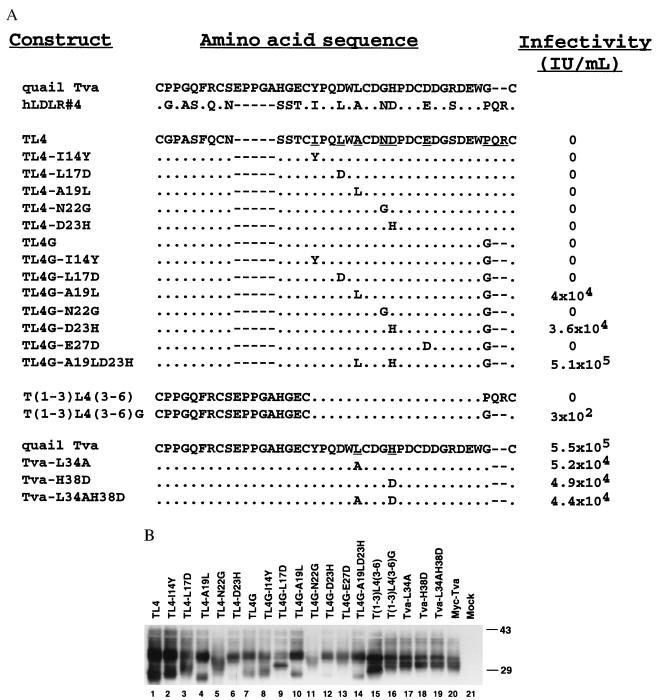
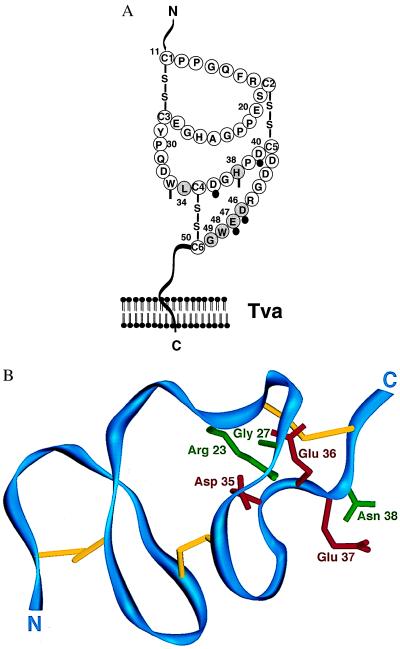
Figure 1.
(A) A cartoon comparison of the quail Tva LDLR module and human low-density lipoprotein receptor module 4 (hLDL-A4) sequences. The Tva LDLR module is numbered 11 (cysteine 1) to 50 (cysteine 6) according to mature Tva sequence, whereas hLDL-A4 is numbered 1 (cysteine 1) to 37 (cysteine 6). Identical residues are shaded. The variant residues from cysteines 3–6 of the two motifs are labeled. The disulfide bonding pattern shown is based on the patterns determined for human LDLR modules 1, 2, and 5 (16–18). (B) A diagram depicting epitope-tagged Tva (Myc-Tva). Two copies of a myc epitope tag were inserted into the N terminus of Tva as shown. The sequence of the myc tags are underlined, and the inserted residues are boxed with dashed lines. The locations of restriction endonuclease enzyme sites for BamHI and SacII used in the construction of the Tva/hLDL-A4 chimeras are shown.
To create a Tva/hLDL-A4 chimeric receptor, the human LDLR repeat 4-coding region was amplified by PCR, using primers OS233 (5′-CCGGATCCGGTAACGGTTCTTTGTCCCGTTGTGGTCCCGCCAGCTTC-3′) and OS234 (5′-GGCCGCGGGGGTCGCGCTGGTCCCACAGCGCTGCGGCCACTC-3′) and a human LDLR cDNA clone as template (13). The amplified DNA fragment was digested with BamHI and SacII and used to replace the Tva sequence in BamHI/SacII-digested Myc-Tva vector. This Tva/LDL-A4 chimera is called TL4. The other chimeric receptors with amino acid substitutions at various positions were generated by standard PCR-based mutagenesis, using TL4 DNA as the template (12).
Chimera and Mutant Nomenclature.
The following nomenclature is used to designate the Tva/hLDL-A4 chimeric receptors (see Figs. 1 and 2A): (i) T stands for Tva, L4 for human LDLR module A4, thus TL4 is the chimera with L4 replacing the Tva A module. (ii) TL4G, G indicates that the residues P34Q35R36 in L4 are replaced by a glycine. (iii) TL4-L17D denotes the TL4 chimera in which L17 is substituted by an aspartic acid. (iv) T(1–3)L4(3–6), residues of the Tva LDL-A module from cysteines 3 to 6 are replaced by the corresponding residues of L4. T(1–3)L4(3–6)G, the P34Q35R36 sequence of L4 is changed to glycine. (v) Tva-L34A, residue leucine 34 of Tva is changed to alanine.
Figure 2.
Sequences, expression, and function of Tva/hLDL-A4 chimeras and Tva mutants. (A) Sequences of the chimeric receptors and mutants along with analysis of their ability to mediate RSV(A) infection. Alignment of the Tva LDLR module and hLDL-A4 is shown at the top (dot, identical residue; dash, gap). The LDLR module of Tva was replaced with human LDL-A4 to generate TL4. Below the TL4 sequence are the series of mutants that were generated based on this chimera as described in the text. The nine nonconserved residues in TL4 targeted for mutation are underlined. At the bottom is the quail Tva sequence and sequences of three mutations in Tva described in the text. The ability of the chimeras and mutants to mediate RSV(A) infection was assayed by using an RCAS(A)-AP vector and is expressed as the number of positive AP-staining cells per milliliter of viral stock (IU/ml). (B) Transient expression of Tva/hLDL-A4 chimeras and Tva mutants in 293T cells. Lysates from 293T cells expressing Tva/hLDL-A4 chimeras were subjected to SDS/PAGE, and the Western blot was probed with a monoclonal anti-myc antibody.
Analysis of Protein Expression and Western Blotting.
Transient expression of the Tva/LDL-A4 chimeras and Tva mutants in 293T and QT6 cells was accomplished by a modified CaPO4-mediated transfection protocol. Briefly, cells were seeded the previous day to give a confluence of ≈50% on the day of transfection. Four hours before transfection, the cells were fed with 10 ml fresh media. Twenty micrograms of plasmid DNA in 388 μl of water was mixed with 62.5 μl 2 M CaCl2 and 50 μl 10× NTE (8.77 g NaCl/10 ml 1 M Tris⋅HCl, pH 7.4/4 ml 0.25 M EDTA to 100 ml with H2O). The DNA mixture was added dropwise to 500 μl 2× transfection buffer (1 ml 0.5 M Hepes⋅NaOH, pH 7.1/0.9 ml 2 M NaCl/0.2 ml 0.1 M Na2HPO4 to 10 ml final volume with H2O). This DNA/CaPO4 mix was added to the media over cells, then the cells were returned to a 37° 5% CO2 incubator for 4–12 hr, after which the cells were fed with fresh media. Preparation of lysates from the transfected cells, SDS/PAGE, and Western blot analysis of the proteins were as described previously (5) except that an mAb, 9E10 (14), to the myc epitope tag was utilized at a 1:2,000 dilution and was detected with a 1:20,000 dilution of peroxidase-conjugated goat-anti-mouse secondary antibody (Pierce).
Infection Assay.
Tva/hLDLR chimeras were transiently expressed in 293T cells by CaPO4 transfection. Twenty-four hours after transfection, the cells were split into six-well plates for infection and 100-mm plates to obtain cell lysates for protein analysis. To determine the level of susceptibility conferred by a particular chimeric receptor, a series of 10-fold dilutions of RCAS(A)AP, a recombinant RSV-A virus carrying an alkaline phosphatase reporter gene, was used to infect the transfected cells. Alkaline phosphatase-positive cells were detected and enumerated 48 hr posttransfection (5) and expressed as number of AP-positive cells per milliliter of viral stock.
Immunofluorescent Detection of Tva Surface Expression.
myc-tagged Tva and the Tva/hLDLR chimeras were transiently expressed in QT6 cells by CaPO4-mediated transfection as described above. Twenty-four hours posttransfection, expression of wild-type or mutant proteins was detected by immunofluorescence following a protocol described previously (15) by using the mAb 9E10 to the myc epitope tag.
Measurement of EnvA Binding.
The binding of Tva/hLDLR chimeras to EnvA was measured by a blocking ELISA protocol. In short, an epitope-tagged, secreted version of EnvA was used to facilitate capture of the envelope protein. This epitope-tagged RSV-A envelope protein (gD-EnvAHis6) contains a 21-residue tag from the gD glycoprotein of herpes simplex virus type 1 fused to the N terminus of EnvA and has the membrane-spanning domain replaced by a six-histidine tail (11). ELISA plates were coated with an mAb (1D3) specific for the gD epitope (1D3 ascites diluted 1:6,000 in 20 mM Tris⋅Cl, pH 8.5/0.1 M NaCl) washed (0.05% Tween-20 in PBS) three times then blocked (0.05% Tween-20/0.5% gelatin, in PBS) for 20 min at 4°. gD EnvA diluted in block buffer was captured onto the plate (75 μl lysate from gD EnvAHis6 expressing cells per well) at 4° for 1 hr. The plates were washed three times, then various amounts of lysate from cells expressing either wt Tva or Tva/hLDLR chimeric proteins diluted in blocking buffer were incubated for 1 hr at 4°C. The plates were again washed three times, and 5 ng of biotin-labeled sTva was added, incubated at 4° for 1 hr, then washed three times. Bound sTva was detected by using avidin-horseradish peroxidase and 2,2′-azinobis(3-ethlbenzthiazolinesulfonic acid. Plates were read after incubation for various lengths of time at room temperature by using a Molecular Dynamics ELISA reader at 405 nm. The percentage of inhibition was calculated with the formula: % inhibition = 100 − 100(S/T), where S is the test sample and T is the total sTva binding when incubated with lysate from mock-transfected cells. Each experiment was performed in triplicate and repeated several times with consistent results.
RESULTS
LDL-A4 Cannot Replace the LDL-A Module of Tva for Viral Receptor Function.
Sequence comparison of the LDL-A module of Tva with the LDLR-like sequences in GenBank revealed that human LDLR repeat 4 (hLDL-A4) is most closely related to the LDL-A module of Tva. Fig. 1A shows a cartoon comparison of the LDL-A module in Tva with hLDL-A4. Both modules are presented as looped structures constrained by three disulfide bonds between cysteines 1 and 3, 2 and 5, and 4 and 6, which are based on the recently reported disulfide-bonding patterns of hLDLR repeats 1, 2, and 5 (16–18). The LDL-A module of Tva is numbered 11–50 according to the mature Tva sequence (10), whereas LDL-A4 contains 37 residues (numbered from 1 to 37; ref. 19). The carboxyl termini of these motifs are the most conserved region with 15 of 23 identical residues between cysteine 3 and 6 (shown in shade). Three residues (DEW) of Tva previously identified as important determinants for virus receptor function (8, 9, 20) are conserved in LDL-A4 at positions 31–33 near the carboxyl terminus.
Because LDL-A4 is highly homologous to Tva in the region between cysteines 4 and 6 (C4–C6), which is critical for viral receptor activity (8), a chimeric receptor was created in which the entire LDL-A module of Tva was replaced with the LDL-A4 module (TL4, Fig. 2A). TL4 DNA was transfected into 293T cells, and transient expression of the chimeric receptor protein was examined by Western blot analysis by using a monoclonal anti-myc tag antibody. TL4 was highly expressed in 293T cells; however, the chimeric protein displayed a slightly different migration pattern on SDS/PAGE compared with wt Tva (Fig. 2B, compare lanes 1 and 20).
The ability of TL4 to mediate infection of RSV-A was examined by using 293T cells transiently expressing TL4 and a recombinant RSV-A vector carrying an alkaline phosphatase reporter gene (RCAS(A)AP). 293T cells expressing wt Tva were efficiently infected by RCAS(A)AP, whereas no infection was seen with TL4-expressing cells (Fig. 2A). Thus, although the LDL-A4 is highly homologous to the LDL-A module in Tva, it cannot functionally substitute for it.
To rule out the possibility that TL4 did not function because the protein was not expressed on the cell surface, we transiently expressed TL4 and wt Tva in quail QT6 cells by transfection and examined protein expression by immunofluorescence. The more adherent QT6 cells were used instead of 293T cells for these immunofluorescence studies. Cell surface and intracellular expression was readily detected for both Tva and TL4 (data not shown), suggesting that surface accessibility does not account for the inability of TL4 to mediate infection. Although surface expression does not always correlate with proper folding of a protein, it is likely that TL4 chimeric protein folds properly on the cell surface because it is thought that each LDL-A module folds independently (2).
Conversion of TL4 to a Functional Viral Receptor by Amino Acid Substitutions.
That TL4 does not mediate viral infection suggests that other amino acids, in addition to DEW, within the LDL-A module of Tva must be important for ligand recognition and specificity. We therefore systematically substituted the nonconserved residues of LDL-A4 with the corresponding residues from the LDL-A module of Tva individually and in combination in the TL4 backbone, generating 13 additional chimeric receptors (Fig. 2A). All the nonconserved residues between cysteines 3 and 6 of hLDL-A4, except serine 30 (Fig. 1A), were changed to the corresponding residue from Tva. Serine 30 of LDL-A4 was not altered because it was demonstrated previously that the corresponding arginine in Tva could be changed to serine without effecting Tva receptor function (8, 9). Similarly, the region between cysteines 1 and 3 of LDL-A4 was not targeted for mutagenesis because this region can be deleted and mutated in Tva without a dramatic effect on receptor function (9, 21), suggesting it is not required for receptor activity. However, as is discussed below, this region may play a role in ligand recognition.
The TL4 mutants were transiently transfected into 293T cells, and expression was examined by Western blot analysis. All the TL4 mutants were expressed well (Fig. 2B, lanes 2–14). Although there is some variation in the receptor levels of the mutants, all are expressed well above the minimum level required for full viral receptor function by wt Tva (21). The numerous bands observed for wt Tva, TL4, and the TL4 mutants are most likely a result of modification by N- and O-glycosylation, which has been observed previously with wt Tva (H. Wang, D. Chu, K.G., H. E. Varmus, and P.B., unpublished data). However, noticeable differences in the migration patterns of these chimeric proteins were observed on SDS/PAGE, probably because of the effects of specific mutations within each chimeric protein (e.g., compare TL4-A19L and TL4G-A19L, Fig. 2B, lanes 4 and 10). The expression of all the TL4 mutants was examined by immunofluorescence in quail QT6 cells as described above, and all were detected readily on the cell surface (data not shown).
Single substitutions in LDL-A4 did not confer RSV-A receptor function to the TL4 chimeric receptor. However, two chimeric proteins bearing pairs of substitutions, TL4G-A19L and TL4G-D23H, mediate viral infection efficiently (Fig. 2A). Combining all three of these alterations in the TL4 backbone (TL4G-A19LD23H) produces a receptor that functions indistinguishably from wt Tva. These results indicate that the amino acids at positions 19, 23, and 34–36 in LDL-A4 are critical for ligand recognition, whereas the other nonconserved residues in LDL-A4 (I14, L17, N22, and E27) are not required for viral receptor activity.
Effect of Mutations in Tva on Viral Receptor Function.
To address whether the analogous residues of Tva play a role in ligand recognition and receptor function, L34 and H38 of Tva were altered to the corresponding residues of LDL-A4 (Tva-L34A, Tva-H38D, and Tva-L34AH38D). These Tva mutants were expressed at similar levels to wt Tva in 293T cells (Fig. 2B, lanes 17–19) and were detected on the surface of QT6 cells by immunofluorescence (data not shown). Transient expression of these mutants in 293T cells and infection of these cells by RCAS(A)-AP viruses revealed that all three Tva mutants were impaired in viral receptor function by approximately 10-fold compared with wt Tva (Fig. 2A). These results indicate that, as suggested by the chimeric protein data, L34 and H38 in Tva participate in ligand recognition.
Role of the N-Terminal Region of the Tva LDLR Motif in Ligand Recognition.
One explanation for the fact that Tva-L34AH38D is functional as a viral receptor whereas TL4G is completely defective in mediating viral infection is that the region between cysteine 1 and 3 of Tva, although dispensable for receptor function in wt Tva, may affect ligand specificity or recognition. To address the role of the amino-terminal region of Tva in ligand recognition, two chimeric receptors were constructed, containing the region between cysteines 1 and 3 from Tva and the region between cysteines 3 and 6 from hLDL-A4. One chimeric protein retained the LDLR4 sequence at the C terminus of the motif [T(1–3)L4(3–6)] whereas the other has a substitution of PQR to G in LDL-A4 [T(1–3)L4(3–6)G]. These mutants were expressed similar to wt Tva in 293T cells (Fig. 2B, lanes 15 and 16), and surface expression was detected in quail cells (data not shown). T(1–3)L4(3–6)G was able to mediate viral infection at a low level, whereas T(1–3)L4(3–6) was completely defective (Fig. 2A). The ability of the N-terminal region of Tva to convert TL4G to a functional receptor suggests that residues in this region participate in ligand recognition. In addition, these results support the conclusion that converting the three-residue sequence, PQR, to glycine at position 49 is required for viral receptor function.
Analysis of Ligand Binding by the Tva/LDL-A4 Chimeras and Tva Mutants.
The relative binding abilities of the Tva/LDL-A4 chimeric proteins were examined by an ELISA that measures the ability of the Tva/LDL-A4 chimeric proteins, prepared as cell lysates from 293T cells expressing these proteins, to block envelope binding by a purified, labeled, soluble form of Tva (sTva). wt Tva (Myc-Tva) efficiently inhibited sTva binding to the EnvA in a dose-dependent manner plateauing by 1 μl and displaying 80% inhibition at the lowest amount (0.1 μl) of Tva used (Fig. 3). In contrast, a majority of the Tva/hLDL-A4 chimeric proteins did not display detectable EnvA-binding activity when 10 μl of cell lysate containing these proteins was used in this assay (data not shown). The only chimeric proteins that demonstrated an ability to bind EnvA are TL4G-A19L, TL4G-D23H, and TL4G-A19LD23H. All three chimeric receptors bind EnvA in a dose-dependent manner; however, compared with wt Tva all three bind envelope poorly. For example, 10 μl of TL4G-A19LD23H or TL4G-D23H lysates display inhibitory activity similar to 100-fold less wt Tva protein (Fig. 3). TL4G-A19L has even less inhibitory activity and apparently has the weakest EnvA-binding activity. Therefore, although in transiently transfected cells these chimeric receptors function efficiently in mediating viral infection, they have a much lower binding activity than wt Tva.
Figure 3.
Binding activity of the Tva/hLDL-A4 chimeras and Tva mutants. The ability of the chimeras and mutant proteins to bind RSV(A) envelope was measured by ELISA as described in Materials and Methods. Various amounts of lysate from 293T cells expressing the chimeric and mutant proteins were used to block binding of labeled sTva. Binding activity is expressed as % inhibition of labeled sTva binding. Envelope binding by three of the Tva/hLDL-A4 chimeras (TL4G-A19L, TL4G-D23H, and TL4G-A19LD23H) and the Tva mutants is compared with Myc-Tva. Bars indicate the standard deviation of three independent experiments performed in triplicate. Not shown are the other chimeras listed in Fig. 2A that did not display envelope-binding activity in this assay.
Analysis of envelope binding by the Tva mutants containing substitutions at residues corresponding to A19 and D23 in LDLR-A4 also reveals that these residues are important for the EnvA-binding activity of Tva (Fig. 3). Tva-H38D required 10-fold more lysate (1 μl) to achieve a similar level inhibition (80%) compared with wt Tva (0.1 μl), whereas a 50-fold excess of Tva-L34A lysate (5 μl) was required for this level of inhibition. The receptor protein containing both these mutations, Tva-L34AH38D, was further impaired and displayed very low levels of EnvA-binding activity when 10 μl of lysate was used. Furthermore, EnvA binding by Tva-L34AH38D was undetectable when lower amounts of lysate were utilized. Thus, these results with Tva bolster the data obtained with the Tva/LDLR-A4 chimeras, suggesting the requirement for the leucine and histidine residues for efficient ligand recognition.
In general, there is good agreement between the binding and infection data for the Tva/LDL-A4 chimeras and mutants. As mentioned, none of the chimeric proteins that were unable to mediate infection displayed detectable binding activity. However, comparison of the binding and infection data reveals differences for some Tva/LDL-A4 chimeras that function as viral receptors. For example, the TL4G-D23H and TL4G-A19LD23H receptors display a similar binding profile but exhibit more than a 10-fold difference in viral receptor function (see Figs. 2A and 3). Although the cause of these discrepancies is not clear, they might reflect the differential abilities of various Tva/LDL-A4 chimeras to participate in postbinding events required to activate the membrane fusion potential of the viral envelope protein. Alternatively, they might simply reflect differences in the binding kinetics of various LDL-A4 mutants that are evident during infection but not binding because these assays measure different aspects of the receptor–envelope interaction.
DISCUSSION
A single LDL-A module in Tva determines envelope-binding specificity and RSV-A receptor function. This module in Tva shares approximately 50% amino acid sequence identity with the fourth LDL-A module in human LDLR and contains conserved residues found in all LDL-A modules. Substitution of the module in Tva with LDL-A4 abolished envelope binding and viral receptor function indicating that nonconserved residues in the LDL-A module of Tva are important for determining ligand specificity. Here we used a gain of function approach to identify residues in an LDL-A module involved in ligand recognition. Mutational analysis of the Tva-LDL-A4 chimera demonstrated that substitution of three nonconserved residues in LDL-A4 with the corresponding residues from Tva converted LDL-A4 to a module functional for both envelope binding and viral receptor activity. Substitution of the corresponding residues in wt Tva with those from LDL-A4 diminished the ability of Tva to bind envelope and to function as a viral receptor. These results illustrate that the gain of function approach used here can be used to identify amino acids responsible for ligand recognition. Furthermore, minor variations in an LDL-A module dictate the specificity of the protein–protein interactions mediated by the module.
Structures for three different LDL-A modules from human LDLR recently have been solved by using both NMR and x-ray crystallography (16–18). These structures reveal that the 40-aa LDL-A module is stabilized by three disulfide bonds (16–18) and an octahedral calcium cage (18). The six cysteine residues and four acidic residues required for this structure are highly conserved in all LDL-A modules including Tva. The vast majority (21/33) of the mutations in LDL-A modules that cause a loss of function and result in FH (4) alter residues that would affect Ca2+ coordination or disulfide bond formation and thus the folding of the LDL-A modules (22). In general, these FH mutations, and many other point mutations that substitute single amino acids in the seven LDL-A modules, affect presentation of LDLR on the cell surface and diminish apolipoprotein binding (4). Indeed, analysis of individual LDL-A modules directly demonstrates that FH mutations cause folding defects (22, 23). Because the majority of FH mutations affect structure of the LDL-A module, it is difficult to use FH mutations as a model to understand ligand recognition by the LDL-A modules. Instead, identification of residues important in ligand recognition might require a simple system such as Tva, in which positive effects on ligand recognition can be assessed readily.
The residues found to be important for Tva viral receptor function in the gain of function experiments described here include L34, H38, and G49 (Fig. 4A). Unlike many of the FH mutations, these three residues are not conserved in LDL-A modules as would be expected for residues that determine ligand specificity. In addition, analysis of the LDL-A5 structure (18) reveals that the side chains of these amino acids are predicted to be on the surface of the LDL-A module (Fig. 4B), suggesting that substitutions of these residues would not affect protein folding and supporting their role in ligand recognition. Finally, altering these residues in LDL-A4 results in a gain of viral receptor function, supporting the hypothesis that L34, H38, and G49 are directly involved in ligand recognition. An alternative role for G49 is suggested by the fact that this residue can be replaced by alanine with no effect on viral receptor function (8, 9) and by the importance of an adjacent tryptophan residue (see below). Perhaps a small residue is required at this position to allow correct alignment of the adjacent tryptophan residue.
Figure 4.
Amino acids of the Tva LDL-A module critical for viral receptor function. (A) A cartoon of the LDL-A module from Tva. The six residues critical for viral receptor function of Tva are shaded and numbered. Residues of the LDL-A module of Tva corresponding to the residues involved in calcium ion coordination in the hLDL-A5 are indicated. Solid circles denote residues with side chains participating in Ca2+ coordination, and vertical bars indicate the residues with backbone carbonyl oxygens contributing to Ca2+ coordination (18). (B) Mapping residues critical for Tva viral receptor function on the structure of LDL-A5. The backbone of LDL-A5 is shown in blue, and the three disulfide bonds are shown in yellow from an angle similar to that in figure 2 of Fass et al. (18). This structure was generated by the program setor using the coordinates of hLDL-A5 deposited in the Protein Data Bank at Brookhaven National Laboratory. The six residues of hLDL-A5 corresponding to those in the Tva LDL-A module (see A) are labeled: green, the side chains of the corresponding residues of Tva identified by this study, Arg-23(Leu-34), Gly-27(His-38), and Asn-38(Gly-49); red, the side chains of the corresponding residues of Tva identified by previous studies (8, 9), Asp-35(Asp-46), Glu-36(Glu-47), and Glu-37(Trp-48). Shown in parentheses are the corresponding residues in Tva.
Previous analysis of mutations that negatively affect Tva viral receptor function suggested that acidic residues 46 and 47 of Tva are important for viral receptor activity (8, 9). These acidic residues are highly conserved in all LDL-A modules and are predicted to form part of the octahedral “calcium cage” required for LDL-A module folding (see Fig. 4A; refs. 18 and 22). Thus, in contrast to previous suggestions that hypothesize a critical role for these acidic residues in ligand binding (1, 8), we propose that it is unlikely these two acidic residues participate directly in ligand recognition, but, instead, mutations of these residues most likely exert their effects by disrupting LDL-A module folding.
Loss of viral receptor function experiments also suggests that an adjacent aromatic residue (W48) in Tva is important for ligand binding and receptor function (8, 9, 20). Unlike the acidic residues, W48 is not conserved among the LDL-A modules, and from the module structure it is not predicted to be important for protein folding. Similarly, Tva loss of function experiments suggested that mutation of L34 impaired viral receptor function, although the importance of this residue for ligand binding was not appreciated previously (8, 9). Interestingly, in the LDL-A5 structure amino acids E37 and R23, which correspond to W48 and L34 of Tva, are adjacent to one another (Fig. 4B) and together with a highly conserved phenylalanine residue (F16 in Tva) could form a hydrophobic pocket important for ligand binding by Tva. The possible requirement of F16 for ligand binding activity has not been explored; however, the proximity of F16 and other residues from the amino terminus of the module to the ligand-binding region may explain the effect of the region between cysteines 1 and 3 upon Tva receptor function. Further mutational studies or structural analysis of Tva or the Tva/EnvA complex will be required to discern whether this model for ligand recognition by the LDL-A module is correct.
Acknowledgments
We thank Joanne Crane for purified sTva, Julie Huang for assistance with the ELISA experiments, members of Bates’ laboratory for useful discussions and technical assistance during the course of this study, Karen Kozarsky for human LDLR cDNA clone, and Di Xia for generating Fig. 4B. This research is supported by grants to P.B. from the National Institutes of Health (CA63531) and American Heart Association (95015200).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: LDL, low-density lipoprotein; LDLR, LDL receptor; RSV, Rous sarcoma virus; EnvA, RSV-A envelope; wt, wild type.
References
- 1.Esser V, Limbird L E, Brown M S, Goldstein J L, Russell D W. J Biol Chem. 1988;263:13282–13290. [PubMed] [Google Scholar]
- 2.Russell D W, Brown M S, Goldstein J L. J Biol Chem. 1989;264:21682–21688. [PubMed] [Google Scholar]
- 3.Hobbs H H, Russell D W, Brown M S, Goldstein J L. Annu Rev Genet. 1990;24:133–170. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 4.Hobbs H H, Brown M S, Goldstein J L. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 5.Rong L, Bates P. J Virol. 1995;69:4847–4853. doi: 10.1128/jvi.69.8.4847-4853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert J M, Bates P, Varmus H E, White J M. J Virol. 1994;68:5623–5628. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connolly L, Zingler K, Young J A. J Virol. 1994;68:2760–2764. doi: 10.1128/jvi.68.4.2760-2764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zingler K, Belanger C, Peters R, Agard E, Young J A. J Virol. 1995;69:4261–4266. doi: 10.1128/jvi.69.7.4261-4266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rong L, Gendron K, Strohl B, Shenoy R, Wool-Lewis R J, Bates P. J Virol. 1998;72:4552–4559. doi: 10.1128/jvi.72.6.4552-4559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates P, Young J A, Varmus H E. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 11.Rong L, Edinger A, Bates P. J Virol. 1997;71:3458–3465. doi: 10.1128/jvi.71.5.3458-3465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 13.Kozarsky K, Grossman M, Wilson J M. Somatic Cell Mol Genet. 1993;19:449–458. doi: 10.1007/BF01233250. [DOI] [PubMed] [Google Scholar]
- 14.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rena S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J, Guo H, Du J, Peiper S C, et al. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daly N L, Djordjevic J T, Kroon P A, Smith R. Biochemistry. 1995;34:14474–14481. doi: 10.1021/bi00044a025. [DOI] [PubMed] [Google Scholar]
- 17.Daly N L, Scanlon M J, Djordjevic J T, Kroon P A, Smith R. Proc Natl Acad Sci USA. 1995;92:6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fass D, Blacklow S, Kim P S, Berger J M. Nature (London) 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, Davis C G, Brown M S, Schneider W J, Casey M L, Goldstein J L, Russell D W. Cell. 1984;39:27–38. doi: 10.1016/0092-8674(84)90188-0. [DOI] [PubMed] [Google Scholar]
- 20.Zingler K, Young J A T. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belanger C, Zingler K, Young J A T. J Virol. 1995;69:1019–1024. doi: 10.1128/jvi.69.2.1019-1024.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blacklow S, Kim P S. Nat Struct Biol. 1996;3:758–762. doi: 10.1038/nsb0996-758. [DOI] [PubMed] [Google Scholar]
- 23.Djordjevic J T, Bieri S, Smith R, Kroon P A. Eur J Biochem. 1996;239:214–219. doi: 10.1111/j.1432-1033.1996.0214u.x. [DOI] [PubMed] [Google Scholar]