Abstract
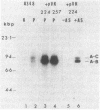
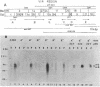
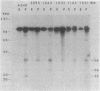
Agrobacterium tumefaciens transfers the T-DNA portion of its Ti plasmid to the nuclear genome of plant cells. Upon cocultivation of A. tumefaciens A348 with regenerating tobacco leaf protoplasts, six distinct single-stranded T-DNA molecules (T strands) were generated in addition to double-stranded T-DNA border cleavages which we have previously reported (K. Veluthambi, R.K. Jayaswal, and S.B. Gelvin, Proc. Natl. Acad. Sci. USA 84:1881-1885, 1987). The T region of an octopine-type Ti plasmid has four border repeats delimiting three T-DNA regions defined as T left (TL), T center (TC), and T right (TR). The six T strands generated upon induction corresponded to the TL, TC, TR, TL + TC, TC + TR, and TL + TC + TR regions, suggesting that the initiation and termination of T-strand synthesis can occur at each of the four borders. Most TL + TC + TR T-strand molecules corresponded to the top T-DNA strand, whereas the other five T strands corresponded to the bottom T-DNA strand. Generation of T strands required the virA, virG, and virD operons. Extra copies of vir genes, harbored on cosmids within derivatives of A. tumefaciens A348, enhanced production of T strands. The presence of right and left border repeats in their native orientation is important for the generation of full-length T strands. When a right border repeat was placed in the opposite orientation, single-stranded T-DNA molecules that corresponded to the top strand were generated. Deletion of overdrive, a sequence that flanks right border repeats and functions as a T-DNA transmission enhancer, reduced the level of T-strand generation. Induction of A. tumefaciens cells by regenerating tobacco protoplasts increased the copy number of the Ti plasmid relative to the bacterial chromosome.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright L. M., Yanofsky M. F., Leroux B., Ma D. Q., Nester E. W. Processing of the T-DNA of Agrobacterium tumefaciens generates border nicks and linear, single-stranded T-DNA. J Bacteriol. 1987 Mar;169(3):1046–1055. doi: 10.1128/jb.169.3.1046-1055.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt-Moerbe J., Rak B., Schröder J. A 3.6-kbp segment from the vir region of Ti plasmids contains genes responsible for border-sequence-directed production of T region circles in E. coli. EMBO J. 1986 Jun;5(6):1129–1135. doi: 10.1002/j.1460-2075.1986.tb04337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. W., Chilton M. D. T-DNA of the Agrobacterium Ti and Ri plasmids. Annu Rev Genet. 1982;16:357–384. doi: 10.1146/annurev.ge.16.120182.002041. [DOI] [PubMed] [Google Scholar]
- Bolton G. W., Nester E. W., Gordon M. P. Plant phenolic compounds induce expression of the Agrobacterium tumefaciens loci needed for virulence. Science. 1986 May 23;232(4753):983–985. doi: 10.1126/science.3085219. [DOI] [PubMed] [Google Scholar]
- Cangelosi G. A., Hung L., Puvanesarajah V., Stacey G., Ozga D. A., Leigh J. A., Nester E. W. Common loci for Agrobacterium tumefaciens and Rhizobium meliloti exopolysaccharide synthesis and their roles in plant interactions. J Bacteriol. 1987 May;169(5):2086–2091. doi: 10.1128/jb.169.5.2086-2091.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C. J., Staneloni R. J., Rubin R. A., Nester E. W. Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol. 1985 Mar;161(3):850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille J., van Kan J., Schilperoort R. trans-Acting virulence functions of the octopine Ti plasmid from Agrobacterium tumefaciens. J Bacteriol. 1984 May;158(2):754–756. doi: 10.1128/jb.158.2.754-756.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinegardner R. T. An improved fluorometric assay for DNA. Anal Biochem. 1971 Jan;39(1):197–201. doi: 10.1016/0003-2697(71)90476-3. [DOI] [PubMed] [Google Scholar]
- Iyer V. N., Klee H. J., Nester E. W. Units of genetic expression in the virulence region of a plant tumor-inducing plasmid of Agrobacterium tumefaciens. Mol Gen Genet. 1982;188(3):418–424. doi: 10.1007/BF00330043. [DOI] [PubMed] [Google Scholar]
- Jayaswal R. K., Veluthambi K., Gelvin S. B., Slightom J. L. Double-stranded cleavage of T-DNA and generation of single-stranded T-DNA molecules in Escherichia coli by a virD-encoded border-specific endonuclease from Agrobacterium tumefaciens. J Bacteriol. 1987 Nov;169(11):5035–5045. doi: 10.1128/jb.169.11.5035-5045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen G. C., Chilton M. D. Activity of T-DNA borders in plant cell transformation by mini-T plasmids. J Bacteriol. 1986 May;166(2):491–499. doi: 10.1128/jb.166.2.491-499.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen G. C., Chilton M. D. The right border region of pTiT37 T-DNA is intrinsically more active than the left border region in promoting T-DNA transformation. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3895–3899. doi: 10.1073/pnas.83.11.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Leroux B., Yanofsky M. F., Winans S. C., Ward J. E., Ziegler S. F., Nester E. W. Characterization of the virA locus of Agrobacterium tumefaciens: a transcriptional regulator and host range determinant. EMBO J. 1987 Apr;6(4):849–856. doi: 10.1002/j.1460-2075.1987.tb04830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A. G. Characterization of nonattaching mutants of Agrobacterium tumefaciens. J Bacteriol. 1987 Jan;169(1):313–323. doi: 10.1128/jb.169.1.313-323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishigaki K., Kaneko Y., Wakuda H., Husimi Y., Tanaka T. Type II restriction endonucleases cleave single-stranded DNAs in general. Nucleic Acids Res. 1985 Aug 26;13(16):5747–5760. doi: 10.1093/nar/13.16.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta E. G., Hellmiss R., Ream W. Overdrive, a T-DNA transmission enhancer on the A. tumefaciens tumour-inducing plasmid. EMBO J. 1986 Jun;5(6):1137–1142. doi: 10.1002/j.1460-2075.1986.tb04338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta E. G., Ream L. W. T-DNA border sequences required for crown gall tumorigenesis. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5112–5116. doi: 10.1073/pnas.82.15.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B., O'Hara P. J., Kwok W., Montoya A. L., Lichtenstein C., Gordon M. P., Nester E. W. DNA from the A6S/2 crown gall tumor contains scrambled Ti-plasmid sequences near its junctions with plant DNA. Cell. 1982 Jul;29(3):1005–1014. doi: 10.1016/0092-8674(82)90464-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Timmerman B., Zambryski P. Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5' virD gene products. EMBO J. 1987 Apr;6(4):857–863. doi: 10.1002/j.1460-2075.1987.tb04831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Zambryski P. C. Agrobacterium tumefaciens and the susceptible plant cell: a novel adaptation of extracellular recognition and DNA conjugation. Cell. 1986 Oct 24;47(2):155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., Zambryski P. C. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell. 1986 Aug 1;46(3):325–333. doi: 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- Thomashow M. F., Karlinsey J. E., Marks J. R., Hurlbert R. E. Identification of a new virulence locus in Agrobacterium tumefaciens that affects polysaccharide composition and plant cell attachment. J Bacteriol. 1987 Jul;169(7):3209–3216. doi: 10.1128/jb.169.7.3209-3216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F., Nutter R., Montoya A. L., Gordon M. P., Nester E. W. Integration and organization of Ti plasmid sequences in crown gall tumors. Cell. 1980 Mar;19(3):729–739. doi: 10.1016/s0092-8674(80)80049-3. [DOI] [PubMed] [Google Scholar]
- Veluthambi K., Jayaswal R. K., Gelvin S. B. Virulence genes A, G, and D mediate the double-stranded border cleavage of T-DNA from the Agrobacterium Ti plasmid. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1881–1885. doi: 10.1073/pnas.84.7.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Herrera-Estrella L., Van Montagu M., Zambryski P. Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from agrobacterium to the plant genome. Cell. 1984 Sep;38(2):455–462. doi: 10.1016/0092-8674(84)90500-2. [DOI] [PubMed] [Google Scholar]
- Wang K., Stachel S. E., Timmerman B., VAN Montagu M., Zambryski P. C. Site-Specific Nick in the T-DNA Border Sequence as a Result of Agrobacterium vir Gene Expression. Science. 1987 Jan 30;235(4788):587–591. doi: 10.1126/science.235.4788.587. [DOI] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Ebert P. R., Stachel S. E., Gordon M. P., Nester E. W. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N. S., Vanderleyden J., Bennett D. R., Barnes W. M., Chilton M. D. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6322–6326. doi: 10.1073/pnas.79.20.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Iwahashi M., Yanofsky M. F., Nester E. W., Takebe I., Machida Y. The promoter proximal region in the virD locus of Agrobacterium tumefaciens is necessary for the plant-inducible circularization of T-DNA. Mol Gen Genet. 1987 Jan;206(1):174–177. doi: 10.1007/BF00326554. [DOI] [PubMed] [Google Scholar]
- Yanofsky M. F., Porter S. G., Young C., Albright L. M., Gordon M. P., Nester E. W. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell. 1986 Nov 7;47(3):471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- Zambryski P., Depicker A., Kruger K., Goodman H. M. Tumor induction by Agrobacterium tumefaciens: analysis of the boundaries of T-DNA. J Mol Appl Genet. 1982;1(4):361–370. [PubMed] [Google Scholar]