Abstract
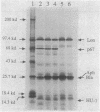
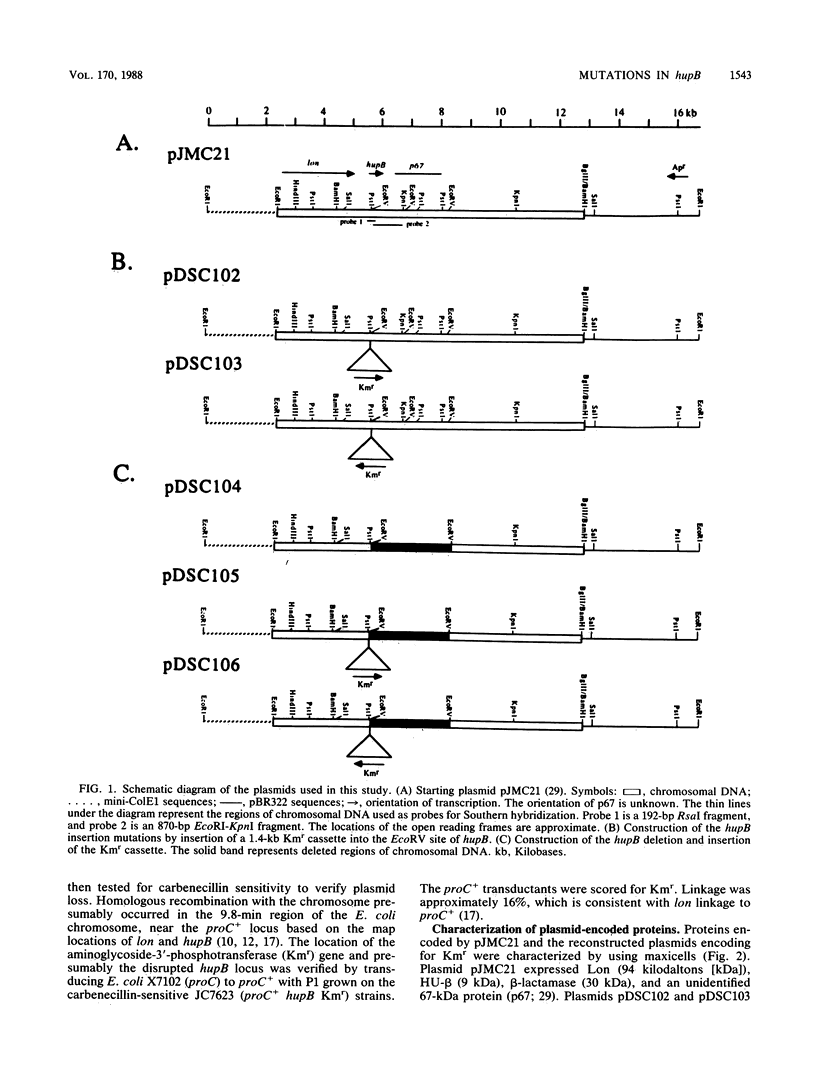
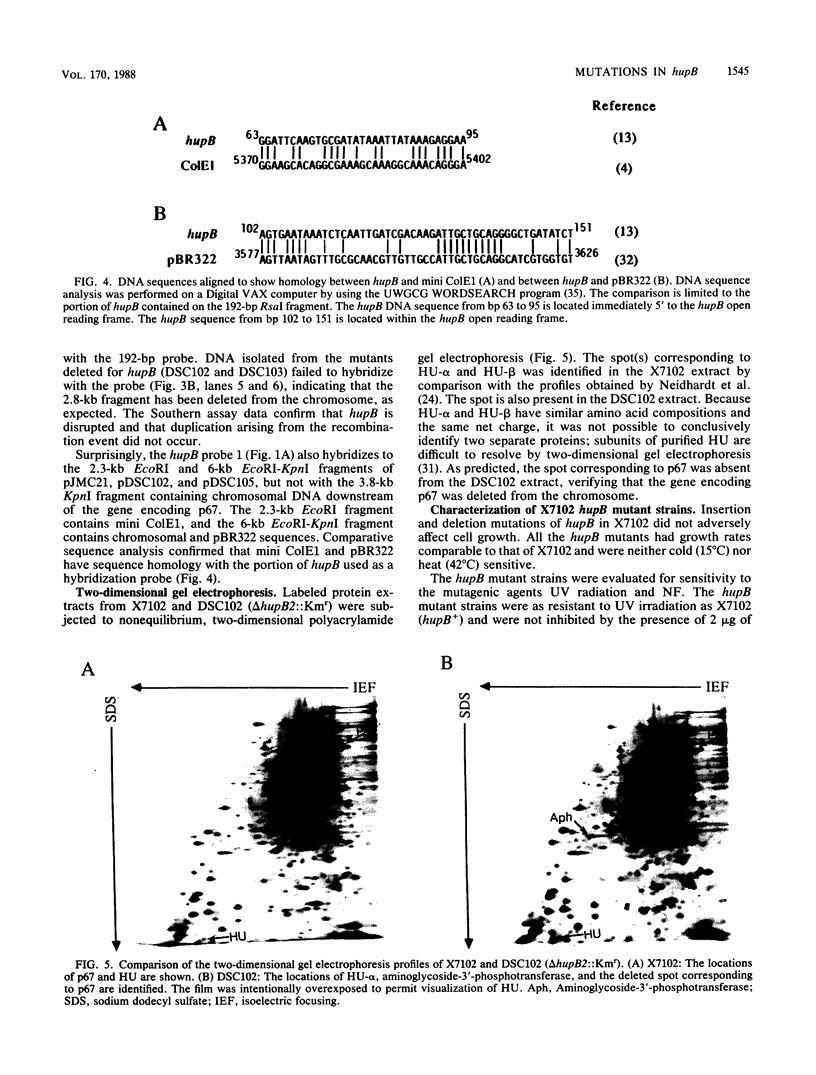
Plasmid pJMC21 contains Escherichia coli chromosomal DNA encoding Lon protease, HU-beta (HU-1), and an unidentified 67,000-dalton protein. A kanamycin resistance cassette was used in the construction of insertion and deletion mutations in hupB, the gene encoding HU-beta on plasmid pJMC21. The reconstructed plasmids were linearized and used to introduce hupB chromosomal mutations into JC7623 (recBC sbcBC). These mutations, as expected, mapped in the 9.8-min region of the E. coli chromosome by P1 transduction (16% linkage to proC+). Southern blot hybridization of chromosomal fragments verified that hupB+ was replaced by the mutant allele, with no indication of gene duplication. All the mutant strains had growth rates identical to that of wild-type E. coli, were resistant to UV irradiation and nitrofurantoin, and supported the in vivo transposition-replication of bacteriophage Mu, Mu lysogenization, Tn10 transposition from lambda 1098, and lambda replication-lysogenization. The only observable phenotypic variation was a reduced Mu plaque size on the hupB mutant strains; however, the yield of bacteriophage Mu in liquid lysates prepared from the mutant strains was indistinguishable from the yield for the wild type.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arps P. J., Winkler M. E. Structural analysis of the Escherichia coli K-12 hisT operon by using a kanamycin resistance cassette. J Bacteriol. 1987 Mar;169(3):1061–1070. doi: 10.1128/jb.169.3.1061-1070.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T., Ohmori H., Tomizawa J., Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985 Jul 25;260(15):8925–8935. [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayda R. C., Stephens P. E., Hewick R., Schoemaker J. M., Dreyer W. J., Markovitz A. Regulatory region of the heat shock-inducible capR (lon) gene: DNA and protein sequences. J Bacteriol. 1985 Apr;162(1):271–275. doi: 10.1128/jb.162.1.271-275.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Elkana Y., Ehrlich S. D., Lederberg J. Electrophoretic separation of Bacillus subtilis genes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2207–2211. doi: 10.1073/pnas.72.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Rudt F., Koch C., Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985 Jul;41(3):771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- Kano Y., Wada M., Nagase T., Imamoto F. Genetic characterization of the gene hupB encoding the HU-1 protein of Escherichia coli. Gene. 1986;45(1):37–44. doi: 10.1016/0378-1119(86)90129-0. [DOI] [PubMed] [Google Scholar]
- Kano Y., Yoshino S., Wada M., Yokoyama K., Nobuhara M., Imamoto F. Molecular cloning and nucleotide sequence of the HU-1 gene of Escherichia coli. Mol Gen Genet. 1985;201(2):360–362. doi: 10.1007/BF00425687. [DOI] [PubMed] [Google Scholar]
- Kirby E. P., Ruff W. L., Goldthwait D. A. Cell division and prophage induction in Escherichia coli: effects of pantoyl lactone and various furan derivatives. J Bacteriol. 1972 Aug;111(2):447–453. doi: 10.1128/jb.111.2.447-453.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARKOVITZ A. REGULATORY MECHANISMS FOR SYNTHESIS OF CAPSULAR POLYSACCHARIDE IN MUCOID MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1964 Feb;51:239–246. doi: 10.1073/pnas.51.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovitz A., Baker B. Suppression of radiation sensitivity and capsular polysaccharide synthesis in Escherichia coli K-12 by ochre suppressors. J Bacteriol. 1967 Aug;94(2):388–395. doi: 10.1128/jb.94.2.388-395.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi M. R., Trisler P., Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985 Dec;164(3):1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I., Nash H. A. Direct role of the himA gene product in phage lambda integration. Nature. 1981 Apr 9;290(5806):523–526. doi: 10.1038/290523a0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Craigie R. Mechanism of bacteriophage mu transposition. Annu Rev Genet. 1986;20:385–429. doi: 10.1146/annurev.ge.20.120186.002125. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Vaughn V., Phillips T. A., Bloch P. L. Gene-protein index of Escherichia coli K-12. Microbiol Rev. 1983 Jun;47(2):231–284. doi: 10.1128/mr.47.2.231-284.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Ross W., Shore S. H., Howe M. M. Mutants of Escherichia coli defective for replicative transposition of bacteriophage Mu. J Bacteriol. 1986 Sep;167(3):905–919. doi: 10.1128/jb.167.3.905-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker J. M., Markovitz A. Identification of the gene lon (capR) product as a 94-kilodalton polypeptide by cloning and deletion analysis. J Bacteriol. 1981 Jul;147(1):46–56. doi: 10.1128/jb.147.1.46-56.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Suryanarayana T., Subramanian A. R. Specific association of two homologous DNA-binding proteins to the native 30-S ribosomal subunits of Escherichia coli. Biochim Biophys Acta. 1978 Sep 27;520(2):342–357. doi: 10.1016/0005-2787(78)90232-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Elledge S. J., Krueger J. H., Walker G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]