Abstract
Telomerase reverse transcriptase (TERT) has been identified as the catalytic subunit of the chromosome end-replicating enzyme in Euplotes, yeasts, and mammals. However, it was not reported among the protein components of purified Tetrahymena telomerase, the first telomerase identified and the most thoroughly studied. It therefore seemed possible that Tetrahymena used an alternative telomerase that lacked a TERT protein. We now report the cloning and sequencing of a Tetrahymena thermophila gene whose encoded protein has the properties expected for a TERT, including large size (133 kDa), basicity (calculated pI = 10.0), and reverse transcriptase sequence motifs with telomerase-specific features. The expression of mRNA from the Tetrahymena TERT gene increases dramatically at 2–5 h after conjugation, preceding de novo addition of telomeres to macronuclear DNA molecules. We also report the cloning and sequencing of the ortholog from Oxytricha trifallax. The Oxytricha macronuclear TERT gene has no introns, whereas that of Tetrahymena has 18 introns. Sequence comparisons reveal a new amino acid sequence motif (CP), conserved among the ciliated protozoan TERTs, and allow refinement of previously identified motifs. A phylogenetic tree of the known TERTs follows the phylogeny of the organisms in which they are found, consistent with an ancient origin rather than recent transposition. The conservation of TERTs among eukaryotes supports the model that telomerase has a conserved core (TERT plus the RNA subunit), with other subunits of the holoenzyme being more variable among species.
The ends of linear eukaryotic chromosomes are protected by DNA–protein complexes known as telomeres. In most species, the DNA portion of telomeres consists of tandem arrays of short repeats (6–10 nt) that are remarkably conserved throughout evolution (1). Telomere shortening (≈50 bp per cell division) has been seen in normal vertebrate somatic cells, in which telomeres are not replicated (2–5). It has been hypothesized that loss of a critical portion of the telomere leads to the onset of cellular senescence in these cells (6, 7). In contrast, the telomeres of unicellular eukaryotes, of germ-line cells of multicellular organisms, and of many human cancer cells do not shorten during cell division (8–10) because of the actions of the ribonucleoprotein enzyme telomerase. Telomerase is a reverse transcriptase that uses an internal RNA moiety as a template for the extension of DNA ends (ref. 11 and reviewed in ref 12). The RNA component of telomerase has been cloned from many different organisms (reviewed in ref. 13).
The protein components of telomerase have proven to be more difficult to identify. The use of ciliated protozoa has facilitated this search because of their extremely large numbers of chromosome ends. Ciliates divide vegetatively in rich medium but will conjugate when starved. During conjugation, the development of a new macronucleus is accompanied by DNA amplification and chromosome fragmentation, resulting in thousands to millions of minichromosomes (reviewed in ref. 14). Telomerase is required for the replication of telomeres during vegetative growth as well as the de novo addition of telomeres to nontelomeric ends during macronuclear development (reviewed in ref. 15).
The first proteins shown to be associated with telomerase were two proteins of 80 and 95 kDa from the ciliate Tetrahymena thermophila, which copurified with enzyme activity and with the telomerase RNA subunit (16). These proteins form a complex that associates with the telomerase RNA and with telomeric substrate DNA (17). They show no strong homology to any previously identified polymerases. A protein (TEP1) related to T. thermophila p80 is associated with telomerase RNA in rat, mouse, and human (18, 19). However, the expression of the human TEP1 does not correlate with telomerase activity in cells and tissues, being ubiquitously expressed (18, 20). The RNA component of human telomerase also is expressed even in cells with no detectable telomerase activity, albeit at lower levels than in immortal telomerase-positive cells (21–23). Together, these data suggest that, at least in human cells, the regulation of telomerase activity is provided by another component.
Another species of ciliate was the source for the biochemical purification of a second family of telomerase proteins that share no homology with those discussed above. An active telomerase RNA–protein complex purified from the ciliate Euplotes aediculatus contained two proteins of 123 and 43 kDa (24). The 123-kDa protein contains reverse transcriptase (RT) motifs (25). Its yeast homolog, Est2p, had been identified independently in a screen for yeast mutants showing telomere length reduction and senescence (26). Genes for homologs of Ea_p123 and Sc_Est2p have been identified in humans, mice, and fission yeast (20, 27–30). Here, we refer to this family of proteins as the TElomerase Reverse Transcriptases (TERTs). Mutation of conserved residues in the RT motifs of TERT resulted in abrogation of telomerase activity in both yeast and human cells (25, 29, 31–33). Inducing expression of the human gene (hTERT) restored telomerase activity to telomerase-negative human cells (32–34). Additional compelling evidence that hTERT is the catalytic protein subunit of human telomerase comes from experiments in which in vitro-translated hTERT and the human telomerase RNA were sufficient to reconstitute telomerase activity (32, 35). It also recently has been demonstrated that ectopic expression of hTERT in normal human diploid fibroblasts is sufficient to extend their lifespan in culture, consistent with a central role of this protein in regulation of telomere length and lifespan in human cells (36). Furthermore, the expression patterns of hTERT mRNA correlate with telomerase activity levels in human cells (20, 27, 28, 33).
We now show that Tetrahymena also has a TERT gene and that its expression is increased greatly during conjugation, a time of new macronuclear telomere formation. We also identify the TERT gene in Oxytricha, a hypotrichous ciliate more closely related to Euplotes and one in which telomeric DNA end-binding proteins have been well studied. These new findings support the conclusion that the TERT catalytic subunit is a universally conserved feature of telomerases.
MATERIALS AND METHODS
Growth of Ciliated Protozoa.
Tetrahymena thermophila strain B7 (from the American Type Culture Collection) was used for cloning of the TERT gene. To study the expression level of TERT, strains CU428.2 and B2086.1 (gifts from Marty Gorovsky, University of Rochester) representing two different mating types were used. Cells were grown vegetatively in 10 mg/ml proteose peptone (Difco) and 0.03 mg/ml sequestrene (CIBA–Geigy) at 30°C with shaking at 100 rpm to a cell density of 1 × 105 cells/ml, then collected by centrifugation for 10 min at 1,000 × g. To initiate starvation, cells grown to a density of 1 × 105 cells/ml were collected by centrifugation and incubated at a density of 3 × 105 cells/ml in 10 mM Tris⋅Cl (pH 7.5) at 30°C without shaking for at least 12 h. Starved cells were induced to conjugate by mixing equal numbers of the two mating types in starvation medium without shaking. Oxytricha trifallax, a gift from David Prescott (University of Colorado), were grown as previously described (37) with Chlorogonium as a food source.
Isolation of T. thermophila RNA and DNA.
RNA was extracted from 108 vegetatively growing cells by using the phenol/guanidinium reagent TriReagent (Molecular Research Centre) following the manufacturer’s instructions. Poly(A)+ RNA was isolated from total cellular RNA by affinity chromatography on oligo(dT) cellulose (New England Biolabs). Genomic DNA was prepared by using a DNA extraction kit (Stratagene).
Cloning of the Tetrahymena TERT Gene and cDNA.
Degenerate oligonucleotides were designed based on the sequences of the E. aediculatus, S. cerevisiae, and S. pombe telomerase RT genes (GenBank accession nos. U95964, U20618, and AF015783, respectively) as follows: primer K231, based on the motif T sequence FFYXTE (forward direction), is 5′-biotin-GCCTATTTYTTYTAYNNNACNGA-3′; K220, based on the motif C sequence DDFL(F/I/L)I (reverse direction), is 5′-CCAGATATNADNARRAARTCRTC-3′; K228, based on the motif 1 sequence R(L/I)(L/I)PKK (forward direction), is 5′-ACAATG(C/A)GNHTNHTNCCNAARAA-3′; K224, based on the motif A sequence CYDSIPR (reverse direction), is 5′-ACGAATC(G/T)NGGDATN(G/C)(T/A)RTCRTARCA-3′; and K227, based on the motif A sequence DIKSCYD (reverse direction), is 5′-CAATTCTCRTARCAN(C/G)(T/A)YTTDATRTC-3′; where D = A, G, or T; H = A, C, or T; N = A, G, C, or T; R = A or G; Y = C or T; and parentheses indicate an equal mixture of the indicated bases. Tetrahymena DNA was amplified by PCR by using primers K231 and K220. To enrich for PCR products that were amplified by both primers (one of which was biotinylated), eight cycles of amplification were carried out, and the product was purified over streptavidin beads. The beads were washed with 0.5× SSC (1× SSC is 0.15 M NaCl/0.015 M Na citrate, pH 7.0). Then the beads were boiled in water to release the product, which was reamplified with K231 and K220. Products in the 800- to 1,100-nt range were gel-purified. A second amplification reaction of Tetrahymena DNA was carried out by using primers K228-K224, and amplification products in the 250- to 350-nt range were gel-purified. To enrich for sequences shared between the two sets of PCR products, K231-K220 products were reamplified, bound to streptavidin beads, and denatured by boiling in water, and the eluate was discarded. Reamplified K228-K224 products were added to the same beads in 0.5× SSC, heated to 94°C, and slowly cooled to 50°C to allow hybridization of common sequences. The beads were then separated from the eluate and washed in 0.5× SSC at 55°C, and the products of hybrid selection were released by boiling in water. The eluted DNA was subjected to semi-nested PCR with K228 and K227. Electrophoresis on a 5% denaturing polyacrylamide gel revealed a prominent 290-nt band that was purified, reamplified, cloned, and sequenced.
DNA surrounding the initial PCR fragments was identified by rapid amplification of cDNA ends [RACE (38)] by using the Marathon cDNA amplification kit (CLONTECH) following the manufacturer’s protocols, using 10 mg of total cellular RNA or 1 mg poly(A)+ RNA for reverse transcription. Because a 5′ Met codon was not identified in the resulting 5′ PCR product, reverse transcription and PCR were repeated by using the SMART kit (CLONTECH), which enriches for full length cDNAs. PCR products were sequenced by using the Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham). Both strands of each template were sequenced, and the sequence was confirmed by using at least two independent PCR products. Genomic DNA also was amplified by using gene-specific primers, and the PCR products were sequenced.
Cloning of the Oxytricha TERT Gene.
O. trifallax genomic DNA was prepared as described (39). A genomic library was generated by digesting the DNA with Mung Bean Nuclease and ligating the resulting blunt-ended fragments into the SmaI restriction enzyme site of the plasmid vector pCRscript SK+ (Stratagene) in the presence of restriction enzyme SrfI (25). A probe for colony hybridization of this library was generated by initially carrying out PCR amplification by using degenerate primers based on motifs B’ and C of the Ea_p123 gene (5′-DGTDATNARRTARTCRTC and 5′-YARACHAARGGHATYCCHYARGG). The 3′ terminus of the Ot_TERT gene then was amplified by using nested PCR with two primers derived from this PCR product and another based on the O. trifallax telomere sequence (T4G4)4. Part of this sequence (nucleotides 2703–3007 of the Ot_TERT gene) was used as a probe for library screening. The clone identified was sequenced on both strands by using an ABI 377 automated sequencer (Perkin–Elmer).
Sequence Analysis.
Multiple sequence alignments were carried out by using the program clustal w (1.60), with gap penalties of 10 (for pairwise alignment) or 50 (for multiple sequence alignment). All other parameters were default. The amino acid similarities shown in Fig. 1B and Table 1 are those assigned by clustal, using the Dayhoff PAM 250 matrix. The phylogenetic tree was constructed by first aligning the motif sequences T, 1, 2 and A–E in clustal. The tree was constructed by using the Neighbor Joining method (40) and was drawn by the program njplot.
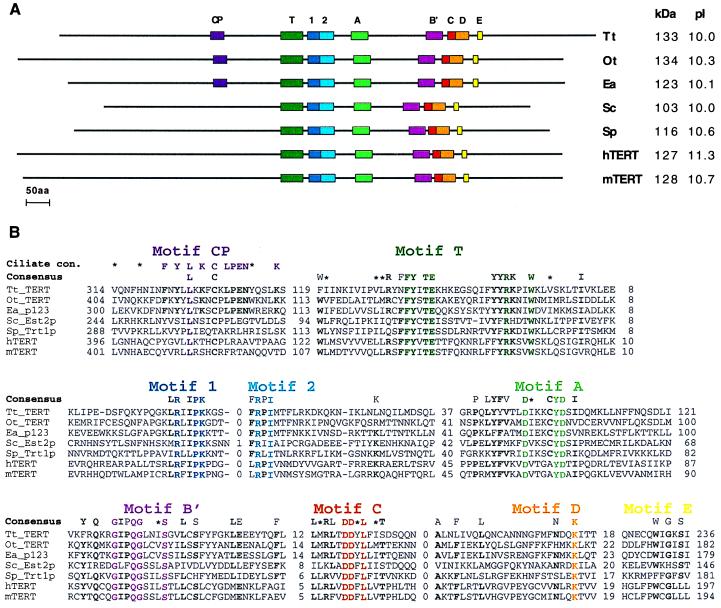
Figure 1.
Primary structure of the seven known telomerase RTs. Organisms represented are Tetrahymena thermophila (Tt_TERT; this paper), Oxytricha trifallax (Ot_TERT; this paper), Euplotes aediculatus [Ea_p123; (25)], Saccharomyces cerevisiae [Sc_Est2p; (26)], Schizosaccharomyces pombe [Sp_Trt1p; (20)], Homo sapiens [hTERT; (20)], and Mus musculus [mTERT; (30)]. (A) Colored boxes indicate the locations of RT motifs 1, 2 and A to E (41), telomerase-specific motif T (20), and the new motif found in ciliated protozoa, CP. kDa, molecular mass in kilodaltons; pI, isoelectric point. (B) Multiple sequence alignment of the motif amino acid sequences. Distances (in amino acids) between motifs and to the ends of the protein are shown. A consensus derived from all seven sequences is shown above them. Amino acids are included in the consensus if they appear in at least five sequences and are typed in bold if the remaining amino acids are conservative substitutions. Colored residues are conserved throughout all seven sequences. ∗, Amino acid similarity. A consensus derived from just the three ciliated protozoa sequences also is shown for motif CP. Abbreviations for the amino acids are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. The nucleotide sequences of the T. thermophila and O. trifallax genes have been submitted to GenBank (accession nos. AF062652 and AF060230, respectively).
Table 1.
Amino acid sequence identity between seven known telomerase reverse transcriptases
| Tt_TERT | Ot_TERT | Ea_p123 | Sc_Est2p | Sp_Trt1p | hTERT | |
|---|---|---|---|---|---|---|
| mTERT | 27 (48) | 25 (49) | 25 (49) | 23 (46) | 31 (49) | 76 (90) |
| hTERT | 25 (47) | 28 (49) | 28 (49) | 26 (46) | 30 (49) | |
| Sp_Trt1p | 29 (47) | 28 (48) | 28 (48) | 30 (48) | ||
| Sc_Est2p | 24 (43) | 25 (46) | 26 (47) | |||
| Ea_p123 | 45 (66) | 63 (79) | ||||
| Ot_TERT | 43 (67) |
Each value is % identity (% similarity in brackets) of motifs T, 1, 2, and A to E, aligned as shown in Fig. 1B.
RT-PCR Analysis of RNA Levels.
Tetrahymena RNA was digested with RQ1 RNase-free DNase (Promega), extracted twice with phenol-chloroform, and ethanol precipitated. RNA was quantitated using UV absorbance at 260 nm. RT reactions were performed with 5 μg of RNA and 0.4 μM random hexamers (Perkin–Elmer) in a 20-μl reaction volume using Superscript II (GIBCO/BRL) following the manufacturer’s protocol. Quantitative PCR was performed to measure the relative abundance of the Tt_TERT mRNA and the telomerase RNA subunit. U1 snRNA was used to normalize the amount of RNA per reaction and the efficiency of the RT reactions; in four experiments, there was no consistent trend of changes in the level of U1 snRNA during starvation or mating. PCR amplification with T5, 5′-AGAAATCTTTTAAATATCTTCGAA-3′, and T3, 5′-CTGATTTTCTTACTTTTTCTCATTGCT-3′, gave a Tt_TERT mRNA-specific PCR product of 246 bp; R5, 5′-ATACCCGCTTAATTCATTCAGATC-3′, and R3, 5′-TTACCACTTATTTGAACCTAATTG-3′, gave a telomerase RNA-specific PCR product of 101 bp; and U5, 5′-CTTACCTGGCTGGAGTTTGCTATC-3′, and U3, 5′-GCGGAGACAGCACTAAGTGCACG-3′, gave a U1 snRNA-specific PCR product of 152 bp. PCR was performed under the following conditions: 1× PCR buffer with magnesium (Boehringer Mannheim); 0.2 μM each dATP, dCTP, dGTP, and dTTP; 1 μM each of the above primer pairs; 67 μCi/ml [α-32P]dCTP; and 0.3 units Taq polymerase in a 15-μl reaction volume. A 5-min denaturation step at 94°C was followed by repeated cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. An appropriate number of cycles was determined experimentally by quantitating the amount of PCR product as a function of cycle number at the highest RNA concentration used and choosing a point within the log-linear range. The U1 and telomerase RNA reactions were done for 13 cycles and the TERT reactions were done for 20 and 25 cycles. The entire reaction mixture then was subjected to electrophoresis in a 5% polyacrylamide gel in Tris borate/EDTA buffer. The gel was dried, and the bands were quantitated by using a phosphorimager (Molecular Dynamics).
RESULTS AND DISCUSSION
Cloning of the TERT Genes.
To determine whether the TERT gene is conserved among the ciliated protozoa, we searched for homologs in T. thermophila and O. trifallax. The cloning method for both genes was based on PCR amplification of genomic DNA by using degenerate primers designed from conserved regions of the RT motifs of the TERT genes known at that time.
In the case of T. thermophila, an initial PCR product of 290 bp was found to encode a peptide sequence with homology to the other known TERTs. Using this sequence, the rest of the gene was obtained by rapid amplification of cDNA ends (RACE), in which short DNA “adaptors” are ligated onto the ends of a library of cDNA molecules and the 5′ and 3′ regions of the gene are amplified with primers based on the sequences of these adaptors and the known portions of the gene. The codon for the N-terminal Met was taken to be an ATG preceded by a TGA “stop” codon 12 bp upstream. The resulting ORF would produce a protein of 1117 amino acids with a calculated molecular mass of 133 kDa.
Amplification of O. trifallax genomic DNA with degenerate primers derived from motifs B’ and C of Ea_p123 gave a PCR product of 174 bp. A longer PCR product was obtained by nested PCR from this sequence to the telomeric sequence present at the ends of all Oxytricha macronuclear genes. A portion of this fragment then was used as a probe in colony hybridization of an O. trifallax genomic library. A full length clone (3642 bp) with telomeric sequences at both ends was identified. Its ORF encoded a predicted protein of 1,132 amino acids (calculated molecular mass = 134 kDa), which shared 63% identity to Ea_p123 across the entire sequence. The two new TERTs are very basic proteins, with calculated pI ≥ 10 like the previously identified TERTs.
Features of the Inferred TERT Proteins.
The primary structures of the predicted proteins encoded by these two genes (named Tt_TERT and Ot_TERT) are shown in Fig. 1A, along with the other members of the TERT family identified to date. Tt_TERT and Ot_TERT proteins contain all seven RT sequence motifs (41) and the telomerase-specific T motif previously identified (20), with motif spacing very similar to that of the E. aediculatus protein. Fig. 1B shows the amino acid sequence alignment and a consensus sequence for all motifs. This consensus refines that previously described (20) with information from three additional TERT proteins, including the recently identified mouse protein (30). Telomerase-specific features of the RT motifs that distinguish them from the retroviral and retrotransposon consensus are now confirmed (20). They include a conserved Arg in motif 1 and an aromatic residue (Tyr or Phe) following the two critical Asp residues in motif C (see Fig. 1B).
We were surprised to discover that the first Phe in the sequence “FFY” of motif T is not conserved in the sequences from T. thermophila and O. trifallax. We subsequently learned, however, that mutation of this Phe to an Ala in hTERT had a minimal effect on telomerase activity, whereas mutations of other conserved residues in this motif greatly reduced activity (32). The two new ciliate sequences thus confirm the boundary of the essential region of the T motif. The function of this motif is as yet unknown.
In addition, we identified a motif (motif CP) specific to the ciliated protozoan proteins, located 113–119 aa upstream of the T motif. This motif shows striking identity among the three ciliate proteins but shows little conservation among the other TERTs. The function of this motif also awaits mutational analysis with recombinant proteins, but it is tempting to speculate that it may be involved in the enormous increase in telomere numbers that is characteristic of the ciliate life cycle.
There is evidence that the amino acid located five residues C-terminal to the first conserved Asp in motif A of DNA polymerases is involved in the discrimination between dNTPs and rNTPs as substrates (reviewed in ref. 42). Reverse transcriptases, which show a strong preference for dNTPs, have only Tyr or Phe at this location (43). The bulky aromatic sidechains of these amino acids are predicted to form a “steric gate” and interfere with the 2′OH of an incoming rNTP. Indeed, replacement of this Phe with a smaller side chain (Val) in the RT of Moloney murine leukemia virus resulted in an almost equal preference for dNTPs and rNTPs in the mutant enzyme (44). All of the TERTs, including that of Tetrahymena, also have a Tyr at this location (see Fig. 1B). Thus, they would be expected to show a strong preference for dNTPs over rNTPs, although Tetrahymena telomerase is able to incorporate rNTPs at low levels (45).
Introns.
Amplification of the Ot_TERT gene in segments of <500 bp from cDNA and genomic DNA resulted in products with no discernible differences in size. Furthermore, the entire gene was sequenced from genomic DNA, and no convincing intronic consensus sequences were found. Thus, we conclude that the Oxytricha gene contains no introns.
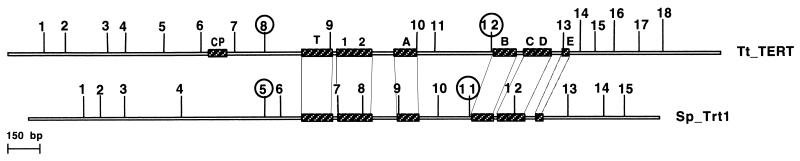
When the Tt_TERT gene was amplified and sequenced from both cDNA and genomic DNA, 18 introns were identified. Each intron had consensus 5′ and 3′ splice site sequences (5′/GTAa… . tAG/3′, where capital and lower case letters indicate bases conserved in 100% and >70% of the 18 introns, respectively). These splice site sequences conform to those previously identified for T. thermophila (46) and resemble those of other organisms. All introns were extremely AT-rich (a mean of 86% with a range of 79 to 92%), as is typical for T. thermophila noncoding regions (46). Their locations are shown in Fig. 2, along with those of the 15 introns identified in the S. pombe gene Trt1, the only other TERT gene for which the intron locations are known. Of interest, two introns show exact matches in location when the proteins are optimally aligned (introns 8 and 12 in Tt_TERT vs. introns 5 and 11 in Sp_Trt1). Other introns also may have concordant locations, although uncertainties in alignment of other regions of the proteins makes this difficult to determine [e.g., introns 11 (Tt) and 10 (Sp)].
Figure 2.
Location of introns in the TERT genes from S. pombe and T. thermophila. Bars represent ORFs with sequence motifs shown as stippled boxes. The two circled pairs of introns show conserved locations in both genes.
The implication is either that the two concordant introns are of relatively early origin, dating at least as far back as the common ancestor of ciliated protozoa and yeast, or that intron insertion is a nonrandom event. If the former is true, then these introns must have been lost in the other organisms for which it is known that there are no TERT introns (O. trifallax, E. aediculatus, and S. cerevisiae). The remaining introns that show no conservation in location must either have moved by “intron sliding” during the evolution of these two lineages or have been inserted more recently. Such intron sliding could occur when a mutation creates a new splice site that preserves an ORF in the adjacent exons.
Phylogenetic Relationships.
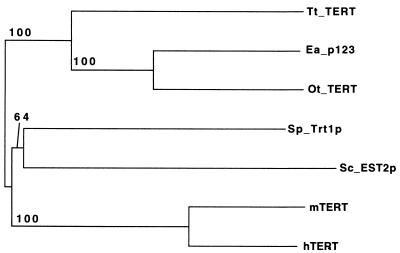
The extent of homology between the motifs of all known TERT proteins and a possible phylogenetic tree based on these relationships are shown in Table 1 and Fig. 3. This tree is consistent with the known evolutionary relationships of these organisms. It is clear that the mouse and human proteins show by far the most homology, as is expected from their relatively recent divergence within mammalian lineages. The ciliate proteins are also more similar to each other than to the other members of the TERT family. Furthermore, the proteins of the two hypotrichous ciliates (Euplotes and Oxytricha) show much greater homology to each other than to the protein of the holotrich Tetrahymena, as expected from their evolutionary relationships. A previously published phylogenetic tree using four of these sequences and only the RT motifs (20) did not recapitulate the known relationships between organisms. Inclusion of additional members of the protein family and an additional motif (motif T) in the analysis thus clarifies the relationship among these proteins.
Figure 3.
A possible phylogenetic tree of all telomerase reverse transcriptases identified to date. The neighbor-joining distance tree was constructed by alignment of motifs T, 1, 2 and A to E as in Fig. 1 and rooted with the ciliated protozoan branch. The statistical support for each node is indicated as the percentage of 1,000 bootstrap replicates that showed that node.
Expression Levels of the Tt_TERT Gene.
Telomerase activity in T. thermophila is greater during macronuclear development than during vegetative growth (47), as would be expected from the massive telomere addition occurring at this time. Activity levels increase ≈1.5-fold upon starvation of the cells and at the peak of macronuclear development (8–10 h after mixing mating types) reach ≈3-fold higher levels than vegetative cells (48). We therefore determined the mRNA expression profile of the Tt_TERT gene.
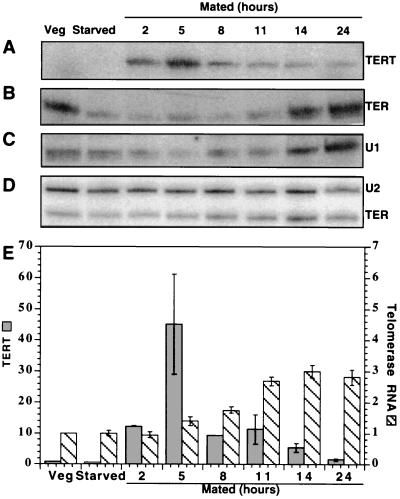
Tt_TERT mRNA was present at very low levels in both vegetative and starved Tetrahymena (Fig. 4A; data not shown). At the first time point (2 h) after mixing different mating types, a significant increase in mRNA was observed, which then peaked 5–8 h after mating was initiated. At that time, 90% of the cells was paired. No increase in mRNA was seen upon continued starvation of a single mating type over this same time period (data not shown). The 45-fold increase in Tt_TERT mRNA level over that in vegetative cells shown in Fig. 4E is most likely an underestimate due to the difficulty in quantitating the low signal in vegetative cells. When the PCR was increased from 20 to 25 cycles, both vegetative and starved cells gave a measurable signal, but the mated samples were reaching the limits of the exponential range for the PCR reaction. Under these conditions, the increase in expression was >100-fold (data not shown). In contrast to the TERT mRNA, the telomerase RNA increased only ≈3-fold during mating (Fig. 4 B, D, and E).
Figure 4.
Expression at the RNA level of T. thermophila telomerase components during vegetative growth, starvation for 14 h, and conjugation for 2–24 h as determined by RT-PCR or Northern hybridization. (A) Tt_TERT mRNA, 20 PCR cycles. (B) Telomerase RNA subunit (TER), 13 PCR cycles. (C) U1 snRNA (49), a control for RNA level and RT-PCR efficiency in the different samples, 13 PCR cycles. (D) Northern hybridization analysis of telomerase RNA, probing for U2 snRNA as an internal standard. (E) Quantitation of TERT mRNA levels (dark columns), divided by the signal obtained for the U1 snRNA and then normalized so that the level in vegetative cells equals 1.0. Each column represents the mean of two separate PCRs, with error bars representing the range of values. Quantitation of telomerase RNA levels as determined by Northern hybridization (striped columns), normalized to U2 snRNA so that the value for vegetative cells equals 1.0. Each column represents the mean of four separate samples, with error bars representing the SEM. (Quantitation of the RT-PCR reaction shown in B gives a similar result, with only the vegetative sample showing a slight difference).
At least superficially, the changes in RNA expression upon induction of telomerase in Tetrahymena resemble those observed in mammalian cells. The human telomerase RNA subunit is present in many telomerase-negative cells but accumulates to higher levels in telomerase-positive cells, whereas the hTERT mRNA levels are much more tightly regulated with respect to telomerase activity (20–23, 27, 28, 33). Thus, it may be that, in Tetrahymena, as in human cells (32–34), the catalytic TERT subunit is a limiting component that can switch telomerase activity on and off.
Conclusions.
Based on in vitro reconstitution experiments, the core catalytic domain of telomerase is hypothesized to consist of the TERT protein in combination with the telomerase RNA (32). The identification of TERT in two more ciliate species supports this concept because it is now clear that TERT is highly phylogenetically conserved. We know that the T. thermophila TERT is expressed coordinately with telomerase activity, and reconstitution experiments confirm that it too is part of the core catalytic domain of telomerase (50). These results support the conclusion that Tetrahymena telomerase, the first such enzyme discovered and the one most extensively studied, has a catalytic subunit related both structurally and evolutionarily to those of other telomerases. The p80 and p95 proteins associated with T. thermophila telomerase RNA (16, 17) now seem more likely to be additional components of a holoenzyme. This conclusion is supported by the lack of p80 and p95 homologs in purified Euplotes telomerase (24) and in the S. cerevisiae genome, although it remains possible that there exist functionally equivalent proteins that differ greatly in sequence. Thus, our results support the concept of a phylogenetically conserved telomerase core, with species variability in the other components of the holoenzyme.
Acknowledgments
We are grateful to Brent Dickinson, Karen Goodrich, Elaine Podell, Yuming Han, Svetlana Gitin, and Barbara Lastelic for technical assistance. We thank David Prescott and Marty Gorovsky for providing us with strains of Oxytricha and Tetrahymena, Toru Nakamura, Joachim Lingner, Art Zaug, and other members of the Cech laboratory for helpful discussions, and Elizabeth Blackburn, Cal Harley and Gregg Morin for comments on the manuscript. This work was supported by Grant GM28039 from the National Institutes of Health.
ABBREVIATIONS
- RT
reverse transcriptase
- TERT
telomerase reverse transcriptase
Footnotes
References
- 1.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 3.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 4.Lindsey J, McGill N I, Lindsey L A, Green D K, Cooke H J. Mutat Res. 1991;256:45–48. doi: 10.1016/0921-8734(91)90032-7. [DOI] [PubMed] [Google Scholar]
- 5.Vaziri H, Schächter F, Uchida I, Wei L, Zhu X, Effros R, Cohen D, Harley C B. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- 6.Harley C B. Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 7.Harley C B, Vaziri H, Counter C M, Allsopp R C. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- 8.Cooke H J, Smith B A. Cold Spring Harbor Symp Quant Biol. 1986;51:213–219. doi: 10.1101/sqb.1986.051.01.026. [DOI] [PubMed] [Google Scholar]
- 9.Allshire R C, Gosden J R, Cross S H, Cranston G, Rout D, Sugawara N, Szostak J W, Fantes P A, Hastie N D. Nature (London) 1988;332:656–659. doi: 10.1038/332656a0. [DOI] [PubMed] [Google Scholar]
- 10.de Lange T, Shiue L, Myers R M, Cox D R, Naylor S L, Killery A M, Varmus H E. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greider C W, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 12.Lingner J, Cech T R. Curr Opin Genet Dev. 1998;8:226–232. doi: 10.1016/s0959-437x(98)80145-7. [DOI] [PubMed] [Google Scholar]
- 13.Greider C W. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 35–68. [Google Scholar]
- 14.Gall J G. Molecular Biology of the Ciliated Protozoa. New York: Academic; 1986. [Google Scholar]
- 15.Blackburn E H. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 193–218. [Google Scholar]
- 16.Collins K, Kobayashi R, Greider C W. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 17.Gandhi L, Collins K. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O Amgen EST Program. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 21.Feng J, Funk W D, Wang S-S, Weinrich S L, Avilion A A, Chiu C-P, Adams R R, Chang E, Allsopp R C, Yu J H, et al. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 22.Avilion A A, Piatyszek M A, Gupta J, Shay J W, Bacchetti S, Greider C W. Cancer Res. 1996;56:645–650. [PubMed] [Google Scholar]
- 23.Bryan T M, Marusic L, Bacchetti S, Namba M, Reddel R R. Hum Mol Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- 24.Lingner J, Cech T R. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 26.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 28.Kilian A, Bowtell D D, Abud H E, Hime G R, Venter D J, Keese P K, Duncan E L, Reddel R R, Jefferson R A. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 29.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S, Mar V, Bass M B, Robinson M O. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenberg R A, Allsopp R C, Chin L, Morin G B, DePinho R A. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 31.Counter C M, Meyerson M, Eaton E N, Weinberg R A. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, et al. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T, Ishikawa F. Nat Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- 34.Counter C M, Meyerson M, Eaton E N, Ellisen L W, Caddle S D, Haber D A, Weinberg R A. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 35.Beattie T L, Zhou W, Robinson M O, Harrington L. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 36.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 37.Swanton M T, Greslin A F, Prescott D M. Chromosoma. 1980;77:203–215. doi: 10.1007/BF00329545. [DOI] [PubMed] [Google Scholar]
- 38.Frohman M A. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 39.Lingner J, Hendrick L L, Cech T R. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joyce C M. Proc Natl Acad Sci USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins K, Greider C W. EMBO J. 1995;14:5422–5432. doi: 10.1002/j.1460-2075.1995.tb00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Csank C, Taylor F M, Martindale D W. Nucleic Acids Res. 1990;18:5133–5141. doi: 10.1093/nar/18.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greider C W, Blackburn E H. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 48.Avilion A A, Harrington L A, Greider C W. Dev Genet. 1992;13:80–86. doi: 10.1002/dvg.1020130113. [DOI] [PubMed] [Google Scholar]
- 49.Ørum H, Nielsen H, Engberg J. J Mol Biol. 1992;227:114–121. doi: 10.1016/0022-2836(92)90686-e. [DOI] [PubMed] [Google Scholar]
- 50.Collins K, Gandhi L. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]