Abstract
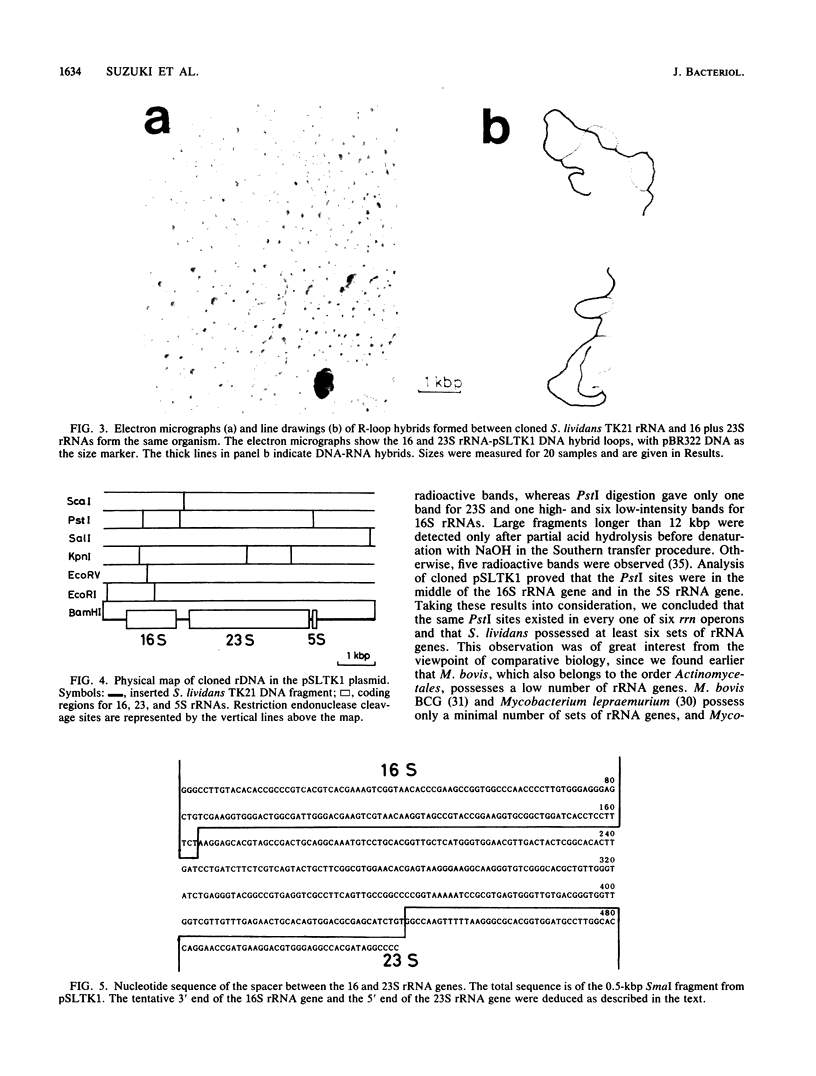
The number of rRNA genes in Streptomyces lividans was examined by Southern hybridization. Randomly labeled 23 and 16S rRNAs were hybridized with BamHI, BglII, PstI, SalI, or XhoI digests of S. lividans TK21 DNA. BamHi, BglII, SalI and XhoI digests yielded six radioactive bands each for the 23 and 16S rRNAs, whereas PstI digests gave one band for the 23S rRNA and one high-intensity band and six low-density bands for the 16S rRNA. The 7.4-kilobase-pair BamHI fragment containing one of the rRNA gene clusters was cloned into plasmid pBR322. The hybrid plasmid, pSLTK1, was characterized by physical mapping, Southern hybridization, and electron microscopic analysis of the R loops formed between pSLTK1 and the 23 and 16S rRNAs. There were at least six rRNA genes in S. lividans TK21. The 16 and 23S rRNA genes were estimated to be about 1.40 and 3.17 kilobase pairs, respectively. The genes for the rRNAs were aligned in the sequence 16S-23S-5S. tRNA genes were not found in the spacer region or in the context of the rRNA genes. The G + C content of the spacer region was calculated to be approximately 58%, in contrast to 73% for the chromosome as a whole.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benigni R., Petrov P. A., Carere A. Estimate of the genome size by renaturation studies in Streptomyces. Appl Microbiol. 1975 Aug;30(2):324–326. doi: 10.1128/am.30.2.324-326.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovier H., Kafri O., Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986 May 14;136(3):1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Bibb M. J., Ward J. M., Cohen S. N. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199(1):26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Boros I., Kiss A., Venetianer P. Physical map of the seven ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1979;6(5):1817–1830. doi: 10.1093/nar/6.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C., Meynell G. G. Rapid screening for plasmid DNA. Mol Gen Genet. 1977 Mar 7;151(2):175–179. doi: 10.1007/BF00338692. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Nomura M. Expression of spacer tRNA genes in ribosomal RNA transcription units carried by hybrid Col E1 plasmids in E. coli. Cell. 1977 Aug;11(4):779–793. doi: 10.1016/0092-8674(77)90291-4. [DOI] [PubMed] [Google Scholar]
- Kenerley M. E., Morgan E. A., Post L., Lindahl L., Nomura M. Characterization of hybrid plasmids carrying individual ribosomal ribonucleic acid transcription units of Escherichia coli. J Bacteriol. 1977 Dec;132(3):931–949. doi: 10.1128/jb.132.3.931-949.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A., Sain B., Venetianer P. The number of rRNA genes in Escherichia coli. FEBS Lett. 1977 Jul 1;79(1):77–79. doi: 10.1016/0014-5793(77)80354-2. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Osawa S. The number of 5 S rRNA genes in Bacillus subtilis. FEBS Lett. 1982 May 17;141(2):161–163. doi: 10.1016/0014-5793(82)80037-9. [DOI] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982 Mar 11;10(5):1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. A., Ikemura T., Nomura M. Identification of spacer tRNA genes in individual ribosomal RNA transcription units of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2710–2714. doi: 10.1073/pnas.74.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neimark H. C. Division of mycoplasmas into subgroups. J Gen Microbiol. 1970 Oct;63(2):249–263. doi: 10.1099/00221287-63-2-249. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome. Nucleic Acids Res. 1983 Sep 24;11(18):6301–6318. doi: 10.1093/nar/11.18.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada M., Muto A., Iwami M., Yamao F., Osawa S. Organization of ribosomal RNA genes in Mycoplasma capricolum. Mol Gen Genet. 1984;196(2):311–316. doi: 10.1007/BF00328064. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart G. C., Wilson F. E., Bott K. F. Detailed physical mapping of the ribosomal RNA genes of Bacillus subtilis. Gene. 1982 Sep;19(2):153–162. doi: 10.1016/0378-1119(82)90001-4. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yoshinaga K., Ono Y., Nagata A., Yamada T. Organization of rRNA genes in Mycobacterium bovis BCG. J Bacteriol. 1987 Feb;169(2):839–843. doi: 10.1128/jb.169.2.839-843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Young R. A., Macklis R., Steitz J. A. Sequence of the 16 S-23 s spacer region in two ribosomal RNA operons of Escherichia coli. J Biol Chem. 1979 May 10;254(9):3264–3271. [PubMed] [Google Scholar]